Microsoft has taken quite an interest in the potential of DNA in computing over the years. Last year Microsoft researchers set a record for DNA data storage. (Its record was beaten this year).
Now Microsoft is turning its attention to the other half of DNA computing, the processor. Researchers at Microsoft have teamed up with scientists at the University of Washington to find a way toward creating super fast computations using DNA molecules.
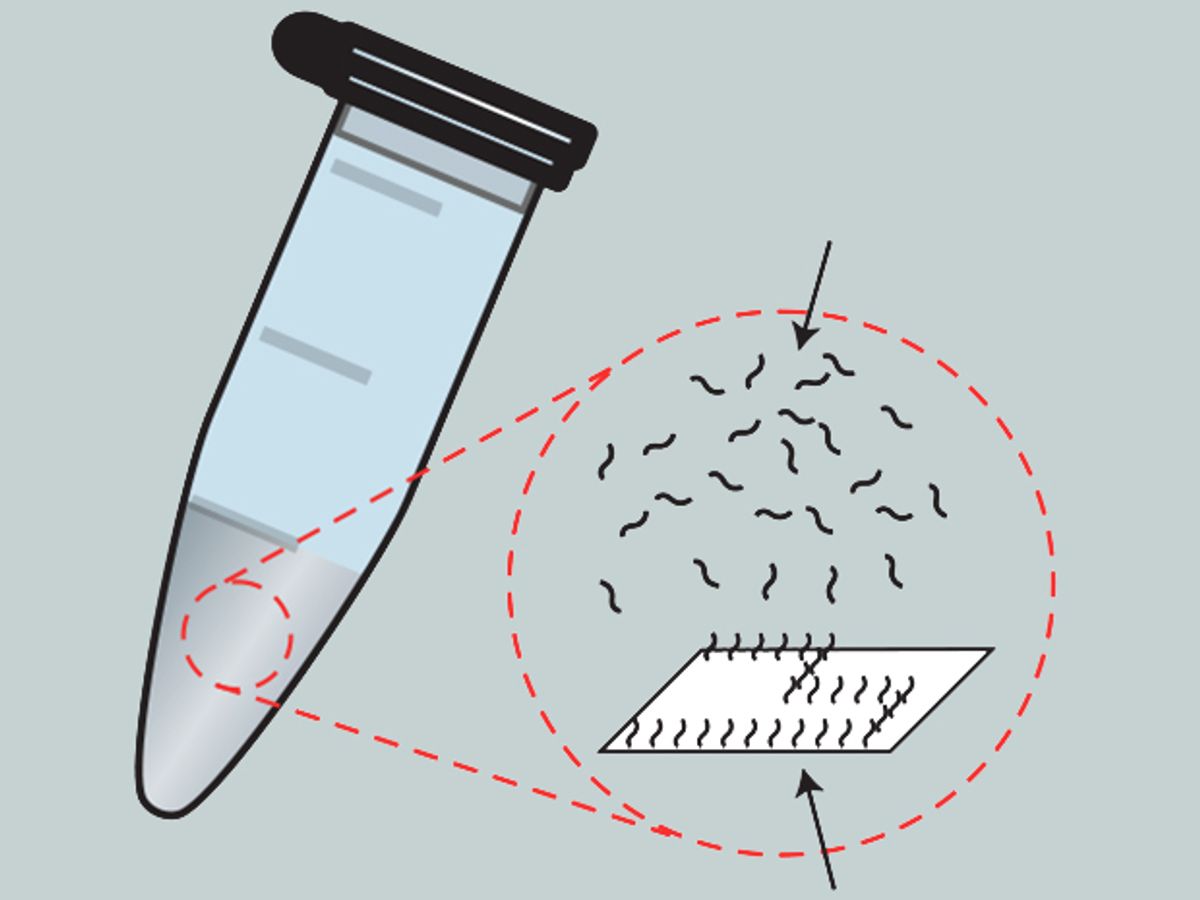
In research described in the journal Nature Nanotechnology, the scientists have developed a method for spatially organizing DNA molecules in regular intervals on a DNA origami surface. That surface is essentially a bunch of DNA strands that have been folded in ways similiar to the techniques of the Japanese art of paper folding. The results offer a new approach to creating DNA logic gates and and the interconnects that link them.
These nanoscale computational circuits are made from synthetic DNA, dubbbed “DNA domino” circuits. These are made from several different strands of synthetic DNA. For example a transmission line consists of hairpin loops of DNA strands with one end afixed to the origami surface. When “input” and “fuel” DNA strands are poured on, they break the loops in the transmission line strands and force them to bend over and link up with their neighbor strand—one after another like dominoes falling—until they form a line of DNA on the substrate.
They used these transmission lines and other structures to make elementary AND and OR gates with two inputs. The researchers were able to to make more complex circuits by linking these elementary gates together.
“The molecular components of the device are spatially positioned in close proximity to one another,” explained Andrew Phillips, the head of biological computation group at Microsoft, in an e-mail interview with IEEE Spectrum. “In our case, the molecular components are DNA strands, and they are fixed in place by attaching them to a DNA origami wafer, which acts as a sort of molecular breadboard.”
In the past, most computational DNA devices consisted mainly of freely-diffusing DNA strands in a chemical soup. Since all of the strands are freely diffusing, they can bump into each other at random and interfere with each other.
“In our case, the components of the devices are positioned close to each other and held in place by a molecular breadboard, such that they are much more likely to interact with their immediate neighbors, and much less likely to interact with other components that are further away, which substantially reduces interference,” said Phillips.
“Our devices do still rely on the presence of a diffusible fuel molecule, so they are not fully-localized, but since most of the components are localized the computation is still significantly faster than a system in which all of the components are feely diffusing,” said Phillips.
All of this close positioning leads to molecular scale computation that is much faster than the relatively slow process of random diffusion—minutes instead of hours. In the research, the scientists measured these devices computing a logical AND using three molecular inputs in seven minutes, compared to four hours for an equivalent DNA circuit with diffusible components.
The production of these devices could be fairly scalable, because they leverage self-assembly, in which the molecules organize themselves. “The devices are designed to be integrated within a DNA breadboard and we rely on the self-assembly of the breadboard to precisely position the interacting DNA components,” said Phillips.
The next step for the researchers will be to investigate how to build larger circuits by increasing the size of the DNA breadboard. This will require advances in DNA origami techniques.
Phillips added: “In addition, we plan to interface these devices with disease biomarkers such as RNA, so that computational logic can be used to accurately diagnose the presence of certain viruses or cancers, initially in blood samples and, ultimately, inside a living cell.”
Dexter Johnson is a contributing editor at IEEE Spectrum, with a focus on nanotechnology.