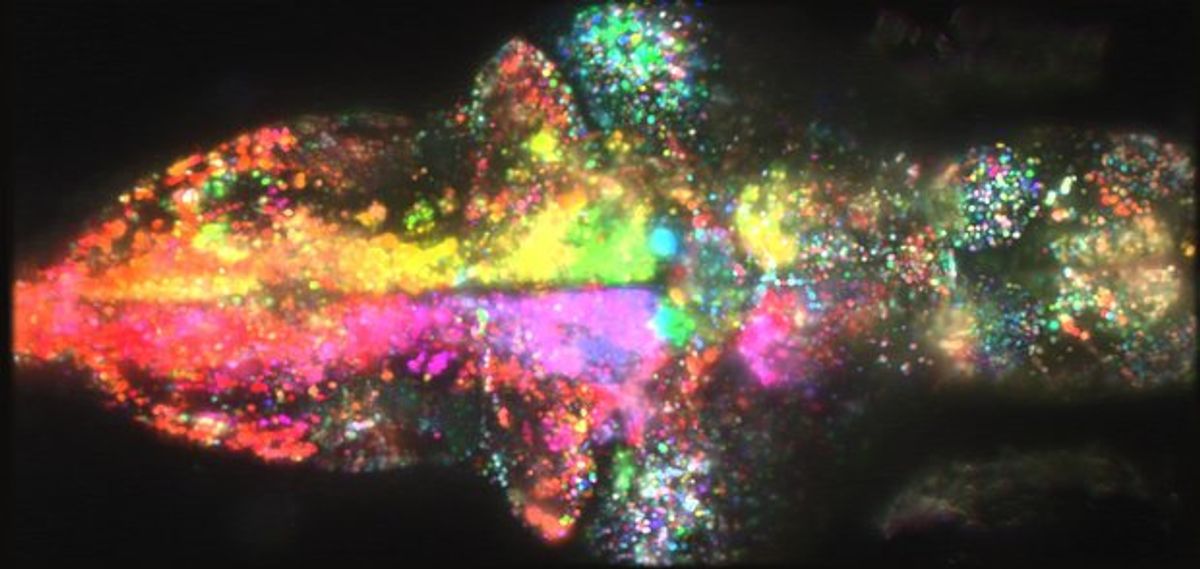
To address this issue, scientists at Janelia Farm Research Campus have come up with a set of analytical tools designed for neuroscience and built on a distributed computing platform called Apache Spark. In their paper in Nature Methods, they demonstrate their system's capabilities by making sense of several enormous data sets. (The image above shows the whole-brain neural activity of a zebrafish larva when it was exposed to a moving visual stimulus; the different colors indicate which neurons activated in response to a movement to the left or right.)
The researchers argue that the Apache Spark platform offers an improvement over a more popular distributed computing model known as Hadoop MapReduce, which was originally based on Google's search engine technology. Here's how Spectrum described these conventional systems in an article on "DNA and the Data Deluge":
While Hadoop and MapReduce are simple by design, their ability to coordinate the activity of many computers makes them powerful. Essentially, they divide a large computational task into small pieces that are distributed to many computers across the network. Those computers perform their jobs (the “map” step), and then communicate with each other to aggregate the results (the “reduce” step). This process can be repeated many times over, and the repetition of computation and aggregation steps quickly produces results.
But the Janelia Farm researchers note that with MapReduce, data has to be loaded from disk for each operation. The Apache Spark advantage lies in its ability to cache data sets and intermediate results in the memory of many computers across the network, allowing for much faster iterative computations. This caching is particularly useful for neural data, which can be analyzed in many different ways, each offering a new view into the brain's structure and function.
The researchers have made their library of analytic tools, which they call Thunder, available to the neuroscience community at large. With U.S. government money pouring into neuroscience research for the new BRAIN Initiative, which emphasizes recording from the brain in unprecedented detail, this computing advance comes just in the nick of time.
Eliza Strickland is a senior editor at IEEE Spectrum, where she covers AI, biomedical engineering, and other topics. She holds a master’s degree in journalism from Columbia University.