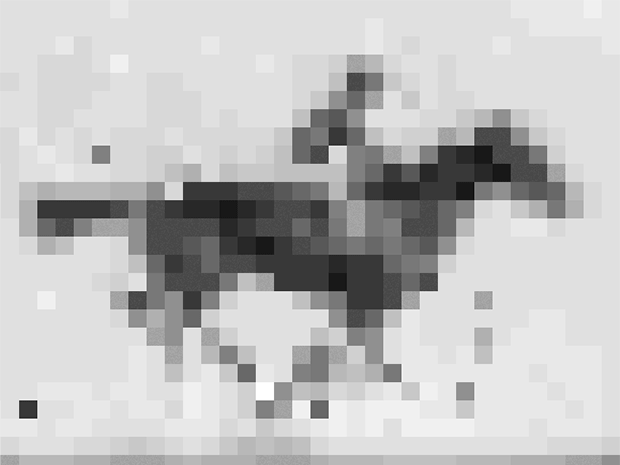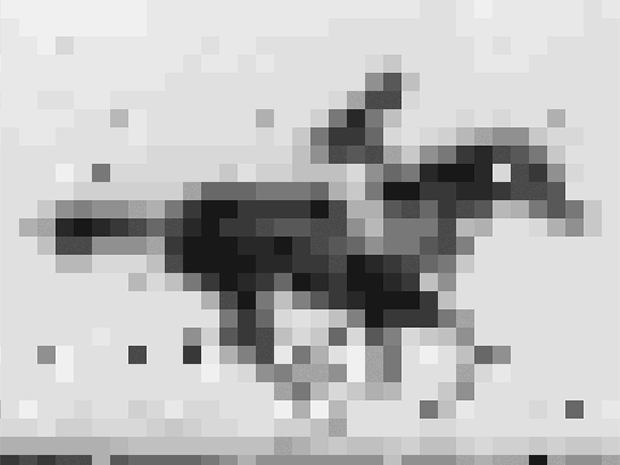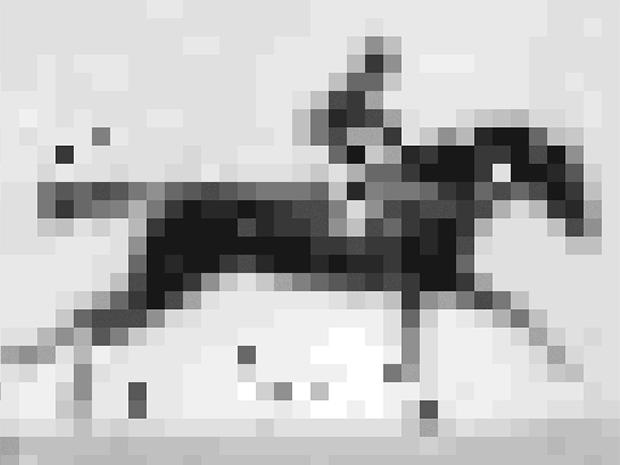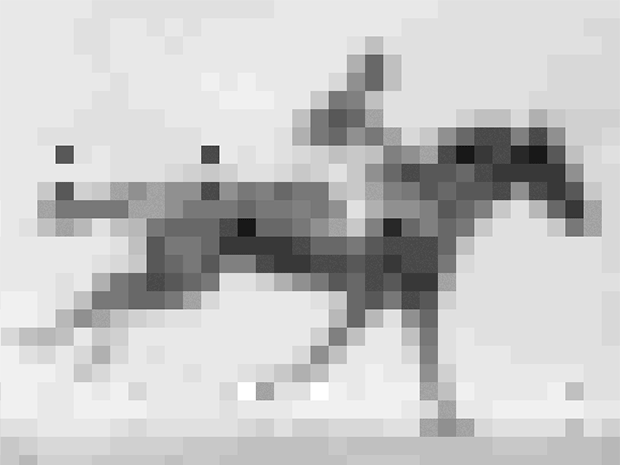The bacteria E. coli might sometimes make us sick, but they have served as a workhorse in science. They teach us about DNA and produce ingredients for drugs and fuel molecules. Now E. coli can claim one more skill: They can store our digital data.
In a report published today in Nature, Harvard researchers demonstrate that it is possible to archive images and movies in the DNA of living E. coli cells.
Researchers are continually developing more efficient ways to store digital data. DNA, the building blocks of life, emerged in the mid-1990s as a potential medium. DNA is, after all, just a code—chemicals symbolized by the letters A, T, G, and C—and it can pack a lot of information into a very small space.
The idea has gained momentum over the last five years. Harvard scientists in 2012 encoded a book in synthesized DNA, and researchers in March reported that they had stored 200 megabytes of data in it—likely the largest amount yet. Even Microsoft has been storing data in DNA. But until now, no one had encoded data right into a living organism, says Seth Shipman, a neuroscientist at Harvard who led the experiments. “That’s much more difficult,” he says.
Shipman and his colleagues encoded into E. coli’s DNA an image and short movie using the gene editing technology CRISPR. The movie, a 36 x 26-pixel GIF of one of the first moving images ever recorded: a galloping mare named Annie G., by Eadweard Muybridge. Shipman and his colleagues retrieved the image and movie with about 90% accuracy using DNA sequencing technology.
So it can be done. But do we want to store our data in living bacteria? Stock our refrigerators with petri dishes full of family memories? (“Mom, where are my prom videos?” “They’re in the E. coli behind the mustard, dear!”)
Perhaps someone will find a use. But Shipman has other plans for his data-storing bacteria: He aims to use it to record the biological activity of cells. “Right now we give DNA information we do know. We want to record information that we don’t know,” he says.
And what we don’t know a lot about is how our own cells in their earliest stages develop. Humans start out as one power-packed ball of pluripotent stem cells, which can turn into anything: brain cells, organ-specific cells, blood cells. But the timing of the development of those cells is not well understood. A data storage system embedded in cells could give us a chronological record of its activity.
But first, Shipman needed to test the system with electronic data. He chose a movie because it allowed him to demonstrate that he could track hundreds of events, in order, over time.
Like many bacteria, E. coli has an excellent internal filing system. A section of its genome, known as CRISPR (clustered, regularly interspaced, short palindromic repeats) runs the show. CRISPR’s job is to grab a piece of DNA from viral invaders and file it in the CRISPR section of its genome, generating a chronological record of invaders.
Shipman’s group imagined that if they made the code for digital information look like that of a virus, E. coli’s CRISPR would be fooled into filing it.
That’s a clever use for CRISPR. Scientists usually use the system as a method for gene editing. Near the CRISPR section of the genome are genes that code for a family of enzymes called Cas, whose job is to slice through DNA strands. Because of that skill, the Cas enzymes—particularly Cas9—have become the tool of choice for scientists who do gene editing. They attach it to a bit of code that can direct it to any spot in organism’s genome, and there it makes a cut, or edit.
Instead of sending out Cas enzymes to chop up distant places in the genome, Shipman’s group let the CRISPR system behave like it normally does: as a mechanism for grabbing viral DNA and bringing it home to the CRISPR section of the genome.
So the group converted the image and movie into short DNA segments that look like fragments of viruses, and, frame by frame, introduced them to the organism. E. coli’s CRISPR system was fooled. It grabbed the fragments and filed them in the order in which they were received. Except instead of getting the code for a virus, E. coli got the code for a movie and an image.
Since E. coli is responsible for at least three foodborne disease outbreaks a year in the US, we feel pretty good about pulling one over on the little suckers.
Emily Waltz is a features editor at Spectrum covering power and energy. Prior to joining the staff in January 2024, Emily spent 18 years as a freelance journalist covering biotechnology, primarily for the Nature research journals and Spectrum. Her work has also appeared in Scientific American, Discover, Outside, and the New York Times. Emily has a master's degree from Columbia University Graduate School of Journalism and an undergraduate degree from Vanderbilt University. With every word she writes, Emily strives to say something true and useful. She posts on Twitter/X @EmWaltz and her portfolio can be found on her website.