Making Chips to Probe Genes
Biochips are now a critical tool for analyzing the human genome--and a lucrative product attracting technology giants
Over a decade of rapid advances in biology has swept an avalanche of genetic information into scientists' laps. But analysis of so vast an input, whether to deduce the inner workings of cells or to diagnose diseases, would be impractical without high-throughput technologies. Of these, DNA microarrays are in the lead. These gene chips or biochips, to use their popular name, allow scientists to look for the presence, productivity, or sequence of thousands of genes at a time. Just five years ago, no practical method could do that.
Analysts predict the US $300 million market in DNA microarrays will leap ahead by about 50 percent per year through 2005. Currently, one company, Affymetrix Inc., Santa Clara, Calif., dominates the market. But as microarrays have caught on among others, Affymetrix has found itself embroiled in patent disputes with a host of other life science firms.
Competition from other technologies may also challenge the company's lead. The potential of various devices for genetics research and drug discovery caught the eye of established high-tech firms like Motorola, Hitachi, Corning, and Agilent Technologies. In often novel ways, each of these firms is adapting existing tools--semiconductors, inkjet printers, flat panel displays--to the manufacture of microarrays, and are developing high-volume manufacturing techniques using fine printing pins and inkjet nozzles to challenge the leading technology, photolithography. Motorola and a few of its rivals are also putting active electronic elements into their microarrays to manipulate and sense DNA.
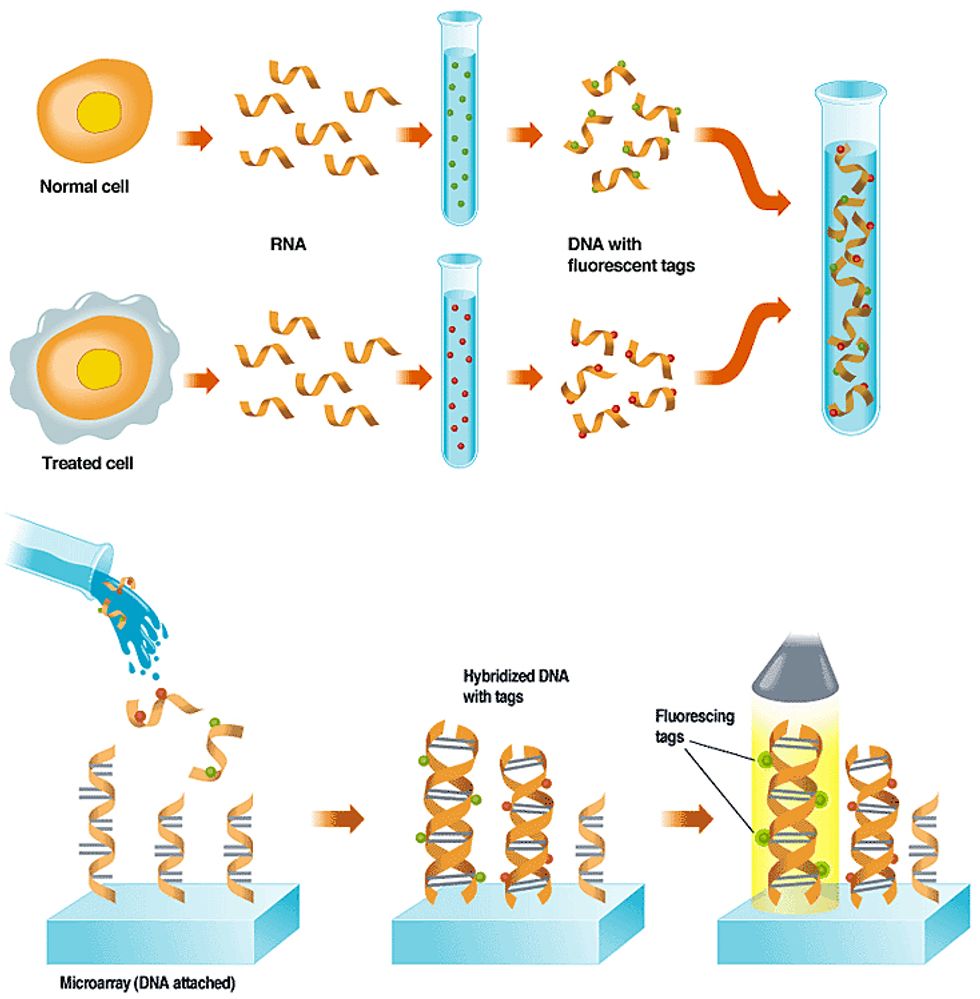
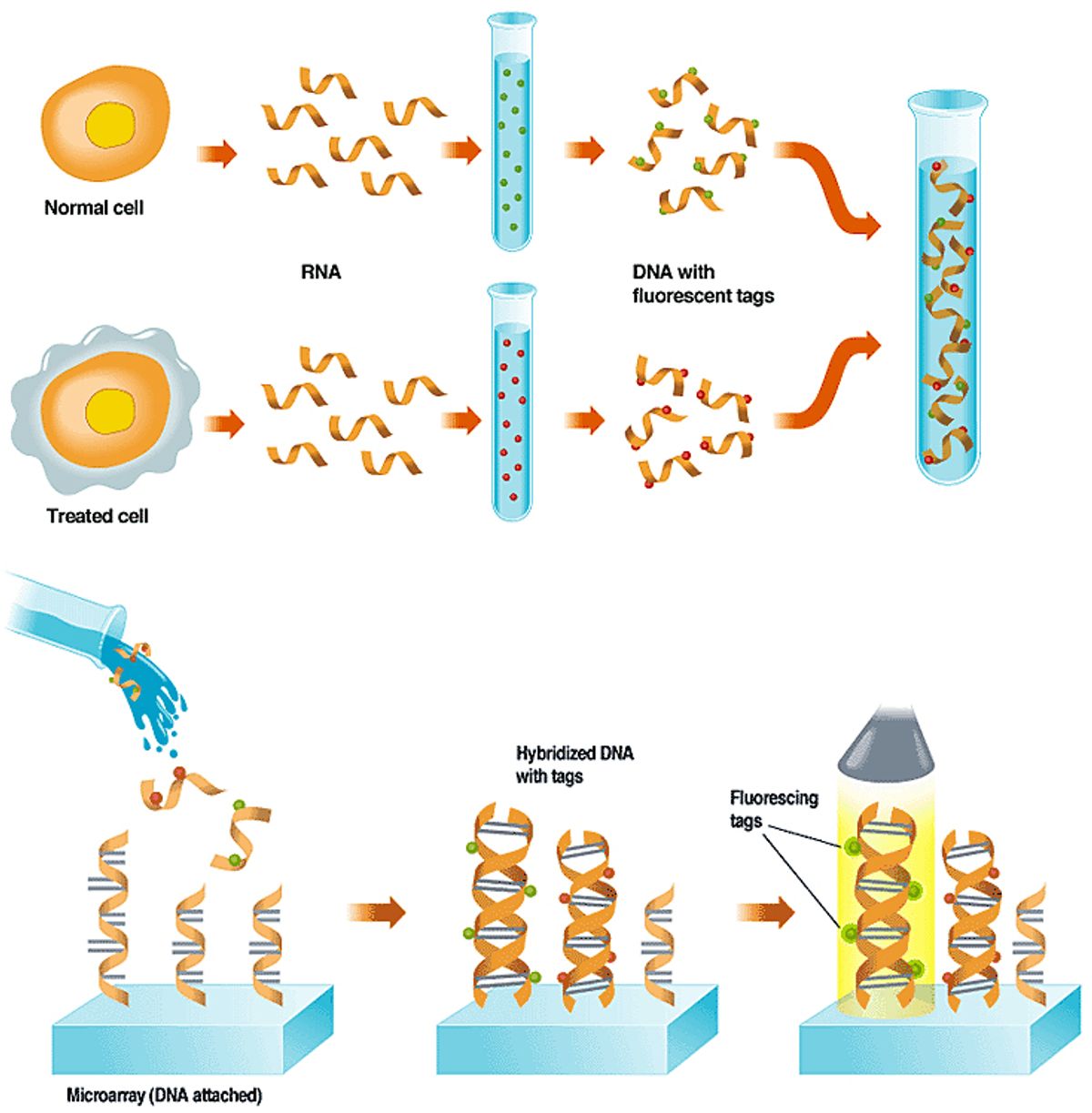
The tagged DNA is washed over a micro-array that has single-stranded DNA fixed to its surface in known locations. This DNA represents important genes or parts of genes. If a gene has been expressed in a cell, it will bind to a copy of itself on the array; those with no complementary site on the array will wash off.
A light source scans the array, causing the dyes to fluoresce. The glow is picked up by a sensor, and a computer interprets the pattern of spots, indicating for each cell which genes are active and the relative abundance of the RNA. Click on the image to enlarge.
Few of these companies are typically identified with the life sciences, and some, like Motorola, are outright newcomers. All are hoping, though, that their technology will compete hard with Affymetrix' and smaller competitors' approaches.
Even with novel technologies, rivals of Affymetrix will need to target its weaknesses or markets it does not serve. "Affymetrix is in a solid position," said Ken Rubenstein, chief executive officer of the Lion Consulting Group in Emeryville, Calif., and author of a recent microarray technology analysis. He expects the company to hold onto most of the research market for the foreseeable future, because of the manufacturing and market infrastructure it has already built up. But there are parts of the market where others might compete. "Agilent is going to be cutting in on the market for customized arrays, and Motorola hopes to make inroads into [low-density arrays], what is now being largely done on a home-brew basis," he said.
The potential profits are huge, because pharmaceutical firms are in a rush to translate the human genome results into new products. First, though, they must figure out what the genes do, how they interact, and how they relate to disease. This is too tall an order for experiments that focus on one gene at a time, but microarrays can perform experiments with thousands of genes simultaneously. Arrays can take snapshots of which subset of genes in a cell is actively making proteins--the term biologists use is "which genes are expressed." Other types of arrays can indicate where mutations lie that might be linked to a disease. Still others could be used to determine if a person's genetic profile would make him or her more or less susceptible to drug side effects.
In 1999, a group of scientists at the Massachusetts Institute of Technology (MIT), Cambridge, performed an experiment that helped establish the importance of microarrays to the research community, particularly to those studying cancer. The MIT group took Affymetrix chips containing 6800 human genes and used them to analyze the expression of genes in cancer cells from two types of blood cancer, namely, acute myeloid leukemia and lymphoblastic leukemia. A standard pathology examination finds it difficult to tell them apart. But the arrays showed a set of 50 genes that have different activity levels in the two cancers, and those genes can now be used to distinguish accurately between the two diseases in patients.
Microarray basics
A microarray starts with a piece of glass, or sometimes a silicon chip, the size of a microscope slide or smaller. Onto this substrate are fixed thousands of patches of single-stranded DNA, called probes, each patch measuring just tens of micrometers across. The location and sequence of each patch of DNA are known ahead of time.
Genes, the instructions for making the proteins that do a cell's work, are encoded in the sequence of chemicals that make up DNA. These chemicals, called nucleotides, each contain a sugar and phosphate backbone plus one of four molecules called bases--adenine, guanine, cytosine, or thymine (A, T, G, C).
In the DNA molecule, the nucleotides are stacked atop each other in two strands forming a twisted ladder. The rungs of the ladder are the bases, with adenine always across from thymine and guanine always across from cytosine; however, under certain conditions the DNA helix can unzip along the rungs of the ladder to form two single strands.
Microarrays draw on the so-called hybridization reaction. Two lengths of single-stranded DNA will bind together, or hybridize, only if the bases on one strand find complementary bases on the other strand. In practice, every adenine base must match up with a thymine, and each guanine with a cytosine.
A leading use of DNA microarrays is in determining which subset of a cell's genes are expressed, or are actively making proteins, under certain conditions, like exposure to a drug or toxic substance. A toxicology experiment, for example, might compare a normal cell to one that had been exposed to a drug [see "Using Arrays to Figure Out which Genes are Turned On,"]. In a diagnostic experiment, such as the leukemia studies described above, two types of cancer cells are used.
The experiment works because when a cell is making protein, the DNA that specifies which type of protein to make is first transcribed into ribonucleic acid, or RNA--a chemical that can encode the same information as DNA. In the lab, scientists extract the RNA and then build single-stranded DNA copies of it. To aid in detecting the DNA on the microarray, they build fluorescent molecules, or tags, into the new DNA.
When the tagged single-stranded DNA is washed over the array, it sticks fast only to any single-stranded probe DNA that has a complementary gene-sequence to its own. A scan of the array with a laser or other excitation source causes any DNA that has found a tagged match to fluoresce, and that glow is picked up by a detector. The detector can consist of a charge-coupled device or a photomultiplier tube.
The image from the detector is then fed to a computer, which analyzes the location, color, and brightness of each patch of DNA. Because the sequence of the array's DNA in each spot is known, the sequence of any DNA captured on that spot is also known. Comparing the colors found at those points on the array reveals the difference in gene expression between the two cells.
Genetic variations called single nucleotide polymorphisms (SNPs) can also be uncovered by microarrays. SNPs are variations or mutations at a single spot in a gene's sequence. Since single-stranded DNA prefers to hybridize only with its perfect complement, arrays can determine the presence of such a mutation. SNPs are thought to be key to why people vary in their susceptibility to diseases.
Experimental arrays containing partial genomes of organisms such as yeast and humans are the bread-and-butter of most firms; but many companies also custom-build arrays from gene sequences that customers upload to them. Most of these sequences for building arrays are available in public databases or from private genomics firms like Incyte Pharmaceuticals Inc., Palo Alto, Calif.
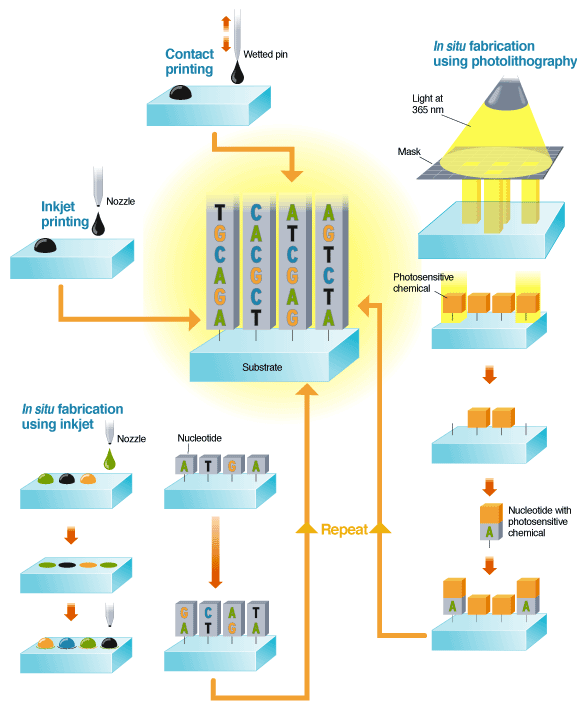
An alternative, known as in situ fabrication, builds the DNA sequence at each site one nucleotide at a time. This is done using either inkjets or photolithography. In inkjetting, solutions of nucleotides are ejected from the nozzle onto the substrate, then chemically fixed to the surface. The next set of nucleotides are jetted onto the first and chemically fixed to those. The process is repeated until the desired set of DNA is complete.
In photolithography, light at 365 nm is shone through a mask to illuminate a subset of regions on a substrate, which is coated with a photosensitive capping chemical. The light releases the capping chemical, exposing parts of the substrate. A solution containing a single type of nucleotide attached to a photosensitive chemical is then washed over the substrate. The nucleotides attach to the unprotected sites, adding their own capping layer. The process is repeated, building up sequences of DNA.
Click on the image to enlarge.
Straight from the wafer fab
There are a number of ways to make arrays, but Affymetrix, the current market leader in microarrays and owner of the now commonly used term "gene chips," may have the one to beat. It makes its high-density arrays with a method familiar to anyone in the semiconductor industry, namely, photo-lithography. This technology, known in the microarray industry as light-directed in situ synthesis, builds DNA probes one base at a time right on the chip [see "Making Microarrays"].
Construction begins with a glass slide that has been chemically primed with sites ready to bind nucleotides. The sites are capped by a photosensitive chemical that detaches under illumination. Light is shone through a patterned mask onto the chip, causing the capping chemical to break away from the areas it strikes, thus exposing the primed spots. A solution containing one of the four types of nucleotides (each molecule of which is itself attached to a capping molecule) is then washed over the chip. The nucleotides bond only to the areas that have been exposed, and add a capping layer themselves. As the process can be repeated with another mask and a different nucleotide, a variety of DNA sequences can be built on the chip.
Photolithography offers the highest density of probes per unit area of any technique in use. Production-scale chips can pack 400 000 probes in 20-µm patches [see "Is There a Moore's Law for Gene Chips?"]. The single-stranded DNA reaches only 25 nucleotides in length, so that it takes several such patches to positively identify a single gene, but Affymetrix claims the redundancy improves detection and quantification of the target gene.
One weakness to the current photolithography method is that a new set of masks must be produced for every new type of array. Help may be in the offing for the latter problem, however. Scientists at the University of Wisconsin in Madison, thee University of Texas Southwestern Medical Center in Dallas, and Xeotron Corp. in Houston have demonstrated a maskless technique that uses an array of micromirrors that reflect onto the appropriate spots on the chip.
Gene spotting
Perhaps the most straightforward array-making method is contact printing. A pin is first dipped into a solution containing pieces of DNA of uniform sequence that have been synthesized in the lab. The pin is pressed to the array surface, leaving behind a droplet of solution [again, see "Making Microarrays"].
Researchers and companies have developed several variations on this basic technique. The most obvious is the replicator pin, whose point must be rewetted after each deposition. Alternatively, pins with a split tip or a hollow tip hold a reservoir of fluid. In a third method, utilized by a recently acquired division of Affymetrix, a pin passes through a ring near its tip before contacting the array surface. The ring, once dipped into a solution of DNA, acts as a reservoir for the pin.
One technology company now moving into microarrays, Corning Inc., of Corning, N.Y., chose the simple replicator pin design but executed it in a novel manner. It prints a thousand spots of DNA simultaneously onto a glass microarray, far more than any other contact printing method.
To break into high-density microarrays, Corning had to develop a system that did not infringe on anybody else's intellectual property (IP). "There is a significant amount of IP in place and we took some time looking at it," said Tom Hinman, division vice president and general manager of Corning's microarray business [see "Transatlantic Scuffle"]. What the company came up with was a printing system that culled technologies from far-flung corners of Corning's own IP storehouse.
For its array, the company cobbled together several technologies it had developed over the years for other purposes. For instance, a three-year-old technique intended for printing color filters onto LCDs was applied to building an array of DNA-printing pins.
That technology involved a type of photosensitive glass into which features on the order of 100 µm can be etched. A pattern is projected onto the glass, which is then doused with hydrofluoric acid. The process yields a print head with 1000 pins about 100 µm in diameter, each separated by 100–120 µm.
Figuring out how to wet each pin with a unique DNA sample was another challenge. Here the company borrowed an extrusion process it had invented in the early 1970s for making porous substrates for catalytic converters. It combined that procedure with one of its techniques for drawing optical fibers, and the result was a funnel-shaped reservoir of 1000 tightly packed conical cells.
To make an array, a high-precision robotic system dips the pin head into the tip of the reservoir and then places it onto a glass slide. Corning currently uses a series of ten 1000-pin heads and reservoirs to produce arrays with 10 000 features each. The company started low-volume production last September, but Hinman told IEEE Spectrum that it will be up to full-scale by the end of this spring.
From inks to nucleic acids
Experience making inkjet printers has been parlayed into a microarray business by Hewlett-Packard spinoff Agilent Technologies Inc., headquartered in Palo Alto, Calif. Inkjetting has two capabilities: it can print spots of DNA sequences synthesized in the lab and also, in a process called in situ fabrication, it can build up parts of genes on the array one base at a time [again, see "Making Microarrays"]. Agilent recently started up a new plant in Santa Clara, Calif., which does both processes.
The inkjet technology used in the Santa Clara plant is essentially the same as that found in a desktop printer. Jets of fluid are pressed through nozzles and broken into uniform droplets by the print head. For in situ synthesis, the four colors of ink--cyan, magenta, yellow, and black--are replaced with nucleotides of DNA having the four types of bases--adenine, guanine, cytosine, and thymine.
This system can build lengths of DNA up to 60 nucleotides long, according to Bill Buffington, vice president and general manager of Agilent's Life Sciences Business. "This is one of those rare cases--and a lot of times this does not happen in nature--where you get both flexibility and low cost at the same time," said Buffington. Of particular help is that different in situ arrays can be synthesized without a change in hardware or chemicals. In contrast, photolithographic methods require a set of masks for each new pattern.
The 60-base length is much shorter than an entire gene, which often runs to hundreds or thousands of bases. But using a proprietary bioinformatics algorithm, Agilent claims that it can take a gene sequence and find a single 60-base sequence within the gene that will effectively identify it for genomics applications.
In contrast to the in situ method, depositing pre-synthesized DNA uses about 100 jet nozzles, each spitting out a unique sequence of DNA. For both methods, the spot size ends up being 70–120 µm in diameter, allowing for arrays with about 25 000 features.
Among the other companies using inkjet spotting to deposit laboratory-synthesized DNA sequences is Motorola Inc. The key difference between its arrays and others is a third dimension. In July 2000 the company inked a $25 million joint development deal with SurModics Inc., Minneapolis, Minn., for making micro-arrays with a coating of acrylimide, a gel commonly used in genetics experiments. Coating a glass substrate creates a three-dimensional jungle of polymer to which DNA can be fixed, as it would be on a flat inkjet array. Motorola claims the gel allows certain enzymatic reactions to occur that might be important to future lab-on-a-chip applications.
In silico
Motorola is also looking at the lower-density-array markets. But many firms have already established a foothold there, and some are blending electronics manufacture and laboratory technology in the way Motorola intends. For example, Nanogen Inc., of San Diego, Calif., has developed a method of targeting DNA to specific sites on a silicon-based array using electronics. In fact, Nanogen and Motorola are engaged in an intellectual property battle over the technology to manipulate DNA on a silicon chip with electric fields.
Instead of making arrays out of glass slides, Nanogen, which has partnered with Tokyo's Hitachi Ltd., produces a chip of silicon embedded with one-hundred 80-µm platinum pads, spaced about 200 µm apart. Each pad can have a voltage of –1.3 to 2.0 V applied to it through external control circuitry. Since DNA carries a negative charge, applying a positive voltage to a site on the chip corrals DNA onto that spot. This can be used to build the array by first washing it in single-stranded probe DNA, biasing the desired spot on the chip, and then chemically fixing the DNA to that spot.
Corning's entry into microarrays relied on a technique initially intended for printing color filters on LCDs
The electronics are also useful during the hybridization reaction, where single strands of DNA find their matches. Pooling DNA onto electrically charged sites speeds the reaction by a factor of as much as 1000, claimed Bob Martinsons, vice president of systems engineering at Nanogen. Conversely, a reverse voltage shakes loose imperfectly matched DNA, leading to more accurate results, he said.
Electric fields might find other uses in microarrays. Motorola is working on a method of detecting hybridized DNA using electrical signals rather than optical ones, Spectrum learned from Nancy Schmelkin, director of marketing for Motorola BioChip Systems. The technology is incompatible with the glass arrays the company is developing because it requires addressable electrodes and other embedded circuitry. So Motorola has begun designing a silicon-based technology to take advantage of its semiconductor manufacturing experience. It plans to combine that experience with technology from the recently acquired Clinical Micro Sensors, Pasadena, Calif., which has developed a process that detects hybridization through a change in conductance.
The future in medicine and research
Nanogen is hoping the improved hybridization speed of its chip will give it a leg up in the medical diagnostics market. That sector is practically nonexistent right now as far as DNA microarrays are concerned, but many in the industry see it as one day becoming the largest market for arrays. For instance, companies envision micro-arrays that can detect the presence of genetic variations that make one drug therapy more efficacious than another.
Some chips are already being tested in clinical laboratories, but estimates of when microarray-aided diagnostics will take off vary. For them to become an important market, the technology will have to improve. In particular, sample preparation time and complexity will have to decrease; so firms expect their current arrays to evolve into more complete on-chip laboratories capable of performing all the necessary procedures to extract genetic material from tissue or blood samples and then analyze it as well.
The research market is shifting as well. As more and more DNA sequences are completed, biologists are looking downstream of DNA for clues to how the body works. And so their attention is turning to understanding proteins and their interactions.
Microarray makers already have their eye on this market. Some start-ups such as Ciphergen Biosystems Inc., of Fremont, Calif., are building their business around arrays of chemicals such as antibodies that will bind and identify proteins much as the DNA devices do. Several firms, including Agilent and Corning, believe their basic microarray platform will be compatible with producing such arrays. And one firm, Packard Instrument Co., of Meriden, Conn., has given up its DNA array business in favor of protein chips. Only time will tell whether protein chips eclipse DNA arrays in importance and whether the big technology firms will succeed in either field.
To Probe Further
Leming Shi, a scientist at BASF Corp. in Princeton, N.J., has cataloged important papers, listed upcoming meetings, and linked to virtually every important DNA microarray Web site at www.gene-chips.com
Microarray Biochip Technology, edited by Mark Schena and published by Eaton Publishing in Natick, Mass., covers many array-making technologies as well as array scanners and bioinformatics software.
Want to learn to make your own microarrays? Try Stanford University Professor Patrick O. Brown's "The Brown Lab's complete guide to microarraying for the molecular biologist" at https://cmgm.stanford.edu/pbrown/mguide/index.html.