For more than 20 years, the practice of using a low-intensity electric current to pull long strands of DNA through nanometer-scale pores in a membrane and measure the electric field variations of the four nucleic acids—A, C, G, T—has been growing as the main approach for DNA sequencers.
We’ve seen the development of this technology reach the point where U.K.-based Oxford Nanopore has been offering portable DNA sequencers based on this fundamental measurement principle for more than a year. Meanwhile, in the research labs, scientists have been tinkering with better materials for the membrane and have started to work with the “wonder material” graphene to see what benefits it might provide in these types of devices.
Now researchers at the National Institute of Standards and Technology (NIST) may have changed the technology paradigm for DNA sequencers in their proposal for an entirely new material architecture that would represent the first DNA sequencer based on sensing motion in the membrane as the DNA thread passes through it.
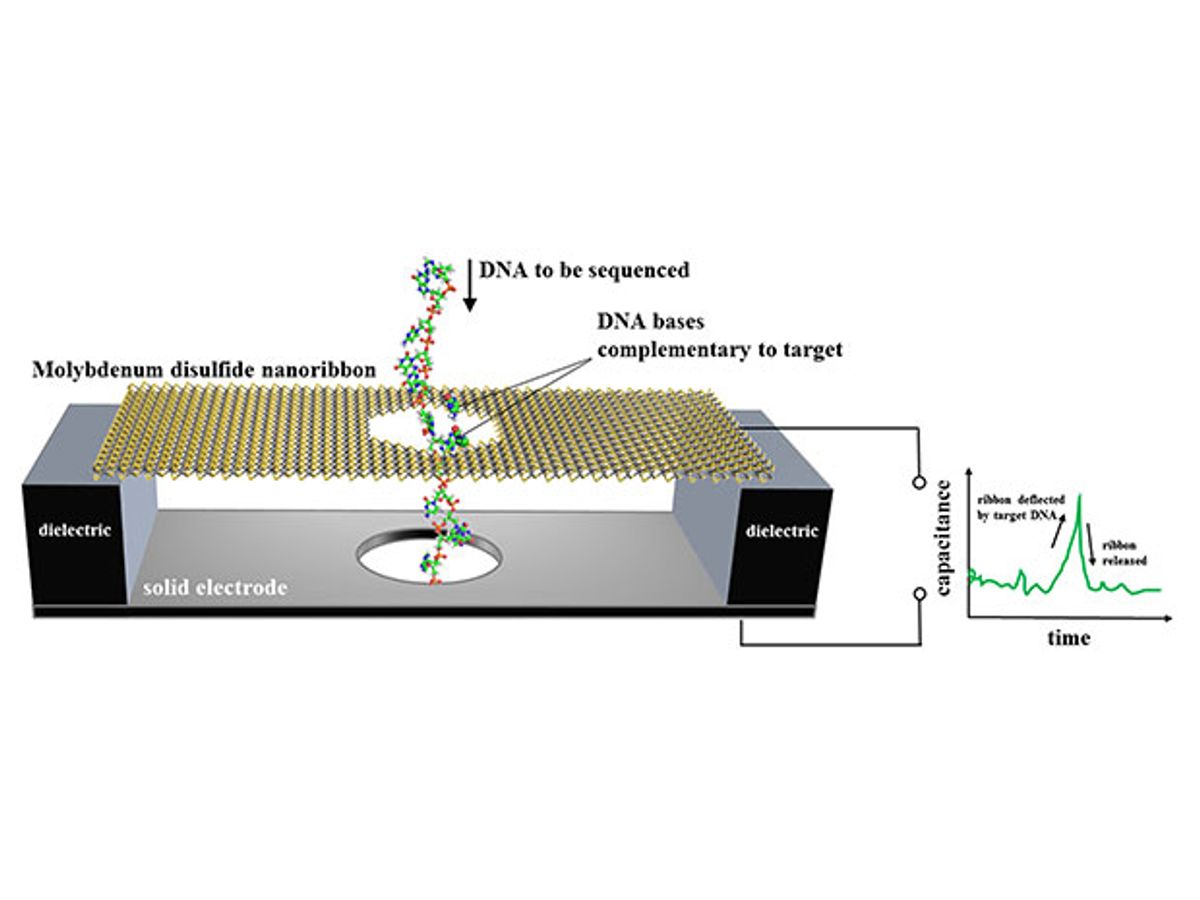
In research described in the journal ACS Nano, the NIST researchers proposed a device in which a nanoscale ribbon of molybdenum disulfide is suspended over a metal electrode immersed in water. In this arrangement, the molybdenum disulfide acts as a kind of capacitor, storing an electrical charge. When a single strand of DNA is passed through a pore in the membrane, the membrane only flexes when a DNA base pairs up with and then separates from a complementary base affixed to the hole. It is this flexing that the motion sensor detects as an electrical signal.
In the paper, the NIST researchers performed numerical simulations of how fast and accurate this DNA sequencer could be, and they concluded that the membrane would be 79 to 86 percent accurate in identifying DNA bases in a single measurement at speeds up to about 70 million bases per second. It is this speed and accuracy that the NIST researchers see as a game changer.
“It is the promise of true single-base resolution and the ability to reliably detect repeated DNA motifs at the rates of millions of bases per second,” said Alex Smolyanitsky, a NIST researcher and lead author, in an email interview with IEEE Spectrum. “An array of sensors described in our paper has the potential to accurately sequence DNA at speeds far greater than anything on the current market, while the device itself is envisioned to be portable and low-power.”
In a head-to-head comparison with research darling du jour graphene, the benefits are clear.
“The molybdenum disulfide is much less prone to ‘sticking’ to DNA, compared to graphene,” said Smolyanitsky. “Also, it is expected to be electrically conductive at room temperature.”
Before a complete prototype is built, the NIST researchers will be working on chemical functionalization of the material. But there does seem to be an urgency to the research with a patent already being sought on the design.
“We have immediate plans and expertise to work on the experimental aspects of this technology,” said Smolyanitsky. “In addition, we are open to forming early-stage partnerships with the industry.”
DNA sequencing may be just be a starting application for this design with a wide variety of nanoelectromechanical system and device applications on the NIST researchers’ horizon.
Dexter Johnson is a contributing editor at IEEE Spectrum, with a focus on nanotechnology.