Germs That Build Circuits
With viruses serving as construction crews and DNA as the blueprint, biotechnology may hold the key to postlithography ICs
By the time you read this, there's a good chance a virus has built a transistor. Last July, a crowd of microbiologists in New York City heard materials scientist Angela Belcher make a bold prediction: within six months, her laboratory at the Massachusetts Institute of Technology (MIT, Cambridge) would have genetically engineered a virus to coat itself in a crystalline semiconductor sheath and locate and bridge two electrodes--thus forming the critical part of a field-effect transistor, the kind on which most computer chips rely. If Belcher delivers, it will dramatically illustrate biology's promise in furthering nanotechnology, the manufacture of circuits and devices only billionths of a meter in size.
Biological self-assembly, as this field of research is called, has a compelling appeal. Living creatures produce the most complex molecular structures known to science. Crafted over eons by natural selection, these three-dimensional arrangements of atoms manifest a precision and fidelity, not to mention a minuteness, far beyond the capabilities of current technology. Under the direction of genes encoded in DNA, cells construct proteins that put together the fine structures necessary for life. And now that scientists can alter the genetic codes of microbes with increasing ease and accuracy, more and more research is showing that this same mechanism can be forced to construct and assemble materials critical not to nature necessarily, but to future generations of electronics.
Belcher's virus gets its circuit-building power from a coat of proteins that interacts at the molecular level with a material to which it's introduced, such as a semiconductor wafer. In projects now under way, scientists are using proteins and DNA, the molecule that encodes genetic data, to construct nanometer-scale crystals of semiconductor atom by atom, bind to precious metals, distinguish between different nanoparticles by their electrical properties, and otherwise choreograph the arrangement of nanoscale components.
Circuits that assemble themselves may sound like wishful thinking. But Tim Gierke, who leads a growing effort at E. I. du Pont de Nemours and Co. (Wilmington, Del.) to apply biotechnology to electronics, says that after three years of research, his company is convinced that biological self-assembly is within the realm of commercial viability.
And the U.S. Army, one of the original sponsors of this field of research, is such a believer that in August it formed the Institute for Collaborative Biotechnologies, a US $50 million research center comprising the University of California at Santa Barbara, MIT, and the California Institute of Technology, to accelerate the work. The Army and others see a role for biological self-assembly in fabricating future sensors, displays, and magnetic storage devices, as well as in energy production and information processing.
But there is no guarantee that this technology, as promising as it is, will really lead to practical nanometer-scale devices. So far, research has been limited to manipulating materials such as crystals of semiconductor, not constructing complete devices. And there are many nonbiological nanocircuit schemes in the works as well.
Most scientists say the technology will first be used to construct sensors consisting of one or a few nanodevices connected to ordinary silicon circuitry. But that's not what drives the research. Their ultimate ambition is to upend current fabrication methods by genetically engineering microbes to build nanoscale circuits based on codes implanted in their DNA. No more cutting patterns into semiconductor wafers, an increasingly arduous process involving lasers, plasma, exotic gases, and high temperatures in expensive industrial environments. Instead, a room-temperature potion of biomolecules will execute, on cue, a genetically programmed chemical dance that ends in a functioning circuit with nanometer-scale dimensions.
"The goal," says Evelyn L. Hu, professor of electrical engineering at the University of California at Santa Barbara (UCSB) and an IEEE Fellow, "is to see if you can generate the next paradigm shift" in electronics.
Evolution is key
Angela Belcher's transistor spawned by a virus will be the result of genetic engineering, but the term is misleading. After all, researchers designed neither the virus nor the proteins that enable it to crystallize semiconductors and bind to metals. (In fact, they're not always sure how they work.) Instead, they discovered these phenomena by unleashing the power of evolution. Just as natural selection engineered the eye, genetic engineers are evolving nanotechnology tools by picking the best molecules for the job from among the variants found in large populations over several generations.
The use of evolution is quite a recent notion. Its potential was first demonstrated only in 1997 by Stanley Brown, a geneticist at the University of Copenhagen in Denmark. He identified peptides--small proteins made of a short chain of amino acids--that can bind gold nanoparticles into visible clumps, much as another protein, the clotting agent fibrin, binds blood cells. Three years later, Belcher, then at the University of Texas at Austin, devised a method to rapidly identify peptides that stick to a slew of valuable semiconductors. The race to exploit biology in electronics was on.
Belcher was already well versed in the organizational power of peptides, having earned her doctorate at UCSB under materials scientist Daniel Morse studying peptides in abalone shell. She figured out how these proteins direct the shell's calcium carbonate to form a protective crystal rather than ordinary chalk and even coaxed them to grow these crystals in the lab.
Then, working with Evelyn Hu, she discovered that peptides could likewise guide the creation of materials such as semiconductors. "Abalone didn't get the chance to evolve an interaction with gallium arsenide or indium phosphide," she says. "The question was whether we could facilitate that kind of interaction ourselves."
Evolving abalone to grow gallium arsenide (GaAs) shells would have been at best a slow process with uncertain prospects for success. Instead, Belcher called up New England Biolabs Inc. (Beverly, Mass.), the Home Depot of biotechnology, and ordered a vial of bacteriophage, a virus that afflicts bacteria, for $300. The variant she used resembles a very fat, rocket-shaped pencil, with a coat of one type of peptide along its painted shaft, another type at the "eraser" end, and a third at the sharpened "point"--each controlled by its own genes. Because they multiply rapidly, bacteriophage evolve and change quickly in the lab when presented with a new challenge, exactly what Belcher needed for the experiments she planned.
The phage Belcher received were engineered to display different seven-part peptides at the eraser ends. All one billion possible variants of the peptide were present in the viral solution. Molecular biologists routinely employ these phage display libraries, as they're called, to identify distinctive peptides--for instance, one that blocks a certain enzyme and might therefore make a useful drug. Belcher used her phage library to target pure crystals of semiconductor, in a process called directed evolution.
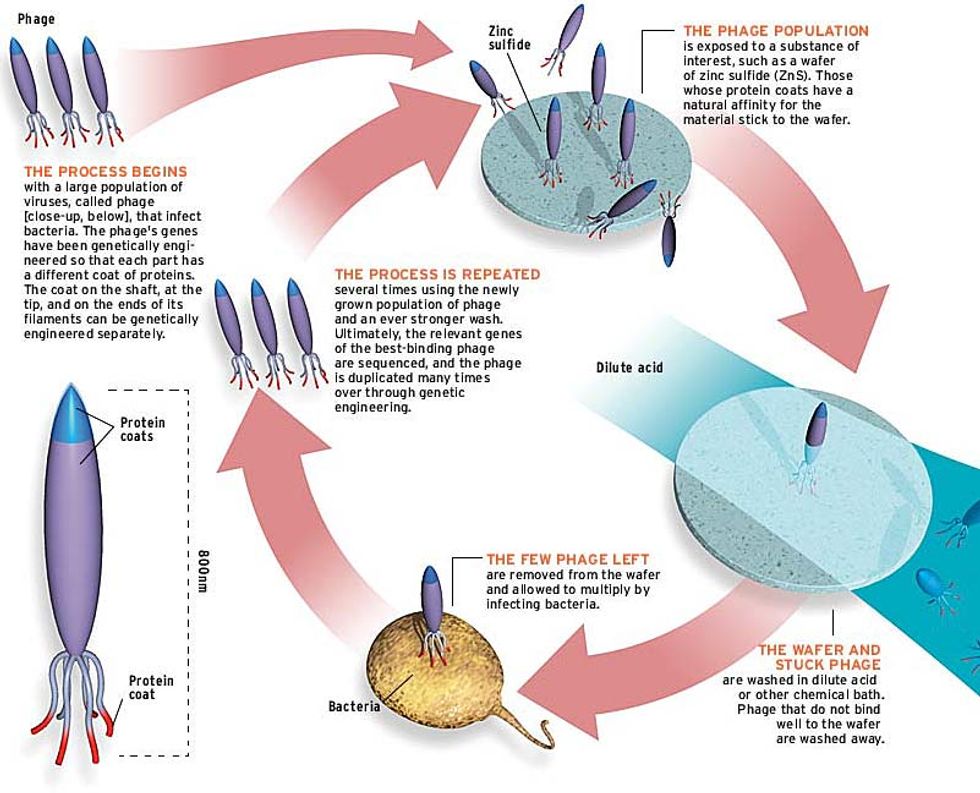
Here's how it works: first, a solution of bacteriophage is poured onto a piece of crystalline semiconductor. Then the chip is rinsed. Most of the viruses are washed away, but a few stick. This sticky subset is removed from the chip, allowed to multiply (by infecting bacteria with them), poured back onto the chip, and then subjected to a more stringent (that is, more acidic or more basic) wash. After several cycles in which the wash becomes stronger and stronger, the viruses sticking to the wafer are those whose peptide coats have a tight fit to the semiconductor crystal [see illustration].
Over the course of a year, Belcher performed rounds of directed evolution to pick out viruses that bind to crystals of gallium arsenide and indium phosphide, two semiconductors employed in high-frequency communications chips. The specificity was incredible: her virus-bound peptides would bear-hug one facet of a GaAs crystal and virtually ignore another.
Belcher and her students have since shown that engineered viruses not only stick to semiconductors but can also be made to form nanometer-scale semiconductor crystals themselves. When mixed with precursor chemicals containing a semiconductor's elemental ingredients, the virus's engineered peptides act as a template, hustling atoms into the same crystal structure to which the peptides were engineered to bind. The result is an organic/inorganic hybrid: viral particles, 7 nm wide and 850 nm long, sporting 2- to 3-nm semiconductor crystals wherever the engineered peptide is found.
These crystals, called quantum dots, are a fundamental component of nanoscale circuits. They have sharp yet tunable bandgaps and have been used to make a plethora of experimental electronic devices, including transistors, solar cells, flash memory, and lasers.
Belcher has selected viruses to form quantum dots of the semiconductors zinc sulfide and cadmium sulfide and the magnetic materials cobalt-platinum and iron-platinum. And she has assembled them into multilayered films, exploiting the fact that viruses will line up shoulder to shoulder in a liquid crystal if the water in which they're dissolved is siphoned off.
Electronics applications for such films aren't hard to imagine. For instance, viral films of magnetic dots could provide the active layer for high-density quantum-dot flash memories. IBM, Fujitsu, and Hitachi, among others, already have such devices in the works. With each quantum dot representing a single bit of memory, a square centimeter of viral film could hold more than 30GB of data.
But circuits require more than just evenly spaced quantum dots. For one thing, they need transistors, which is where genetic engineering again comes in. Once Belcher has found the peptide she's after using directed evolution--one that binds to zinc sulfide or some other semiconducting material--she can identify the genetic code that produces it. Then she can splice that code into the viral DNA wherever she wants--for instance, in the gene that makes proteins along the shaft of the virus rather than at its tail. In the process, she can make the virus form quantum dots all along its peptide coat, essentially forming a tubular 10-nm-wide wire surrounding the virus.
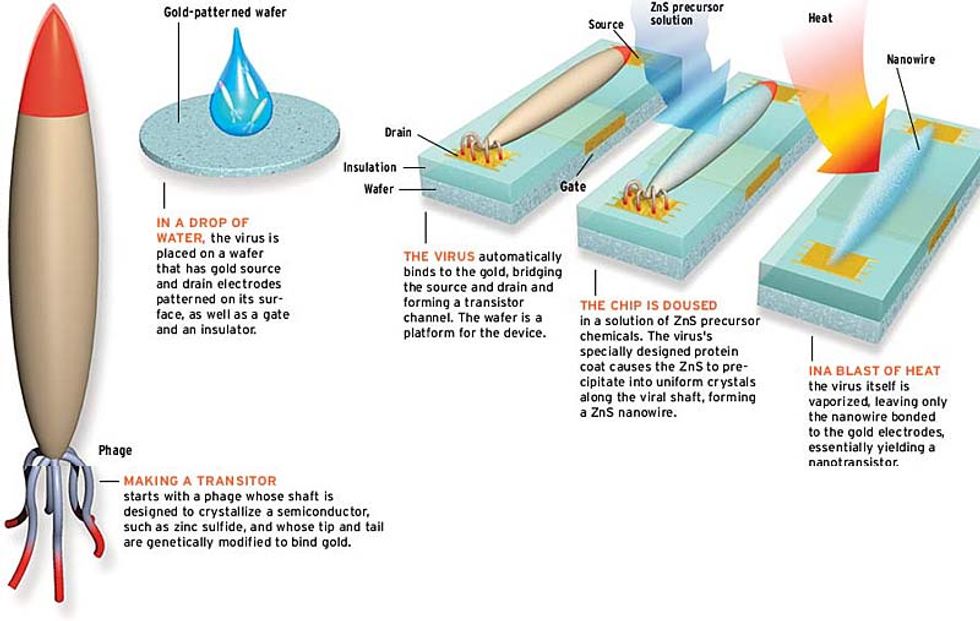
And there's more. Using the same technique, Belcher hopes to make these wired viruses bind to metals such as gold (used for electrodes) by engineering peptides at their tips and tails. At that stage, making a complete transistor requires patterning a silicon wafer so it has exposed source and drain electrodes with a gate electrode between them buried under a layer of insulation [see illustration].
First, the virus would be poured onto the wafer, where its gold-seeking ends would find and bind to the electrodes, making a viral bridge across the gate. Next, semiconductor precursor chemicals would be added, turning the virus's coat into a nanowire. To make a working transistor, it may also be necessary to burn away the virus, leaving only the nanowire attached to the electrodes.
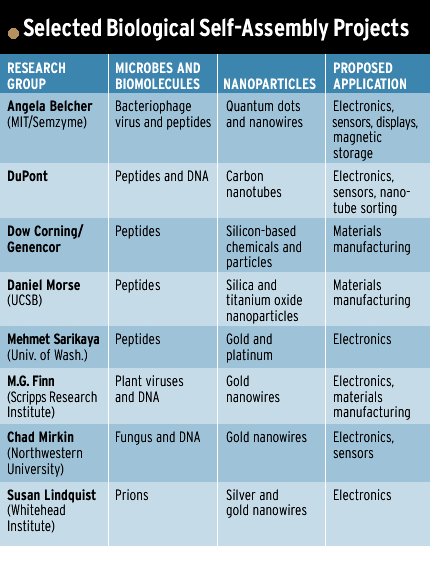
Other self-assembly schemes leave out the viruses, using just their peptides. Some researchers are even exploring naturally derived proteins, such as the mad cow disease-inducing prion [see table].
Mehmet Sarikaya, a materials scientist at the University of Washington, used a technique similar to Belcher's to find peptides that bind to gold and other metals. He thinks these peptides could eventually shuttle the various building blocks of nanocircuits into place. "For these to all assemble at the same time, they have to recognize each other at the molecular scale," he says. And that's a trick peptides can do.
Viralware vendors
In 2001, Belcher and UCSB's Evelyn Hu founded Semzyme (Cambridge, Mass.), a company that will exploit biological self-assembly to make electronic materials as well as more biotechnology-specific applications, such as long-term storage of DNA. The company is set to begin operations this year and is choosing a first product to bring to market.
Big, established companies are taking this research seriously, too. The Army's Institute for Collaborative Biotechnologies has attracted sponsorship from Aerospace Corp., Applied Biosystems, Genencor, IBM, SAIC, and Becton, Dickinson.
Genencor, in particular, took an early interest in bioengineering viruses, forming a $35 million partnership with silicon materials giant Dow Corning in 2001. In the short term, the two firms are merging peptides with silicon-based chemicals to make fabric treatment and cosmetic products. Sensors and other electronics elements are future targets.
DuPont, too, is tinkering with bioevolved peptides. According to Tim Gierke, the company has identified one short-term application: purifying carbon nanotubes. Recently, these hollow pipes just a few nanometers wide have been turned into experimental logic circuits and other devices. Depending on the nanotube's structure, it acts as either a semiconductor or a metal. Unfortunately, current methods generate tubes of both types along with a messy soup of soot, and there's no good way of sorting anything out.
So DuPont evolved peptides that selectively grab the nanotubes and ignore other forms of carbon. To separate the semiconductors from the metallics, the company turned to another important biomolecule--DNA. DuPont scientists discovered that when a particular form of DNA and carbon nanotubes bind, metallic and semiconducting tubes can, to a degree, be separated using a common laboratory trick.
Genetic engineering
Under ordinary conditions, DNA exists as two complementary strands. Individual strands will spontaneously snap together into a tight double-stranded helix, as long as each carries the right sequence of the nucleic acids adenine, cytosine, guanine, and thymine. For instance, a strand containing the sequence ATCG will stick only to its opposite number, TAGC.
In 1996, Northwestern University chemistry professor Chad Mirkin and his colleague Robert Letsinger showed that this behavior could be harnessed to make circuits if DNA were used as a sort of molecular Velcro to organize nanometer-scale structures. By attaching one DNA strand to one set of gold particles and its complement to another, Mirkin and Letsinger got the gold to bunch together. The clumps were neither sophisticated nor valuable, but the technique's potential was clear: when wrapped in several sequences of complementary DNA Velcro, nanocomponents such as nanowires, quantum dots, and carbon nanotubes should spontaneously assemble into complex structures--including circuits.
In fact, Mirkin's lab has come up with a technique to program such self-assembly. Now being commercialized by NanoInk, a Chicago-based start-up, dip pen nanolithography (DPN) employs the tip of an atomic force microscope to deposit "inks" containing various dissolved compounds.
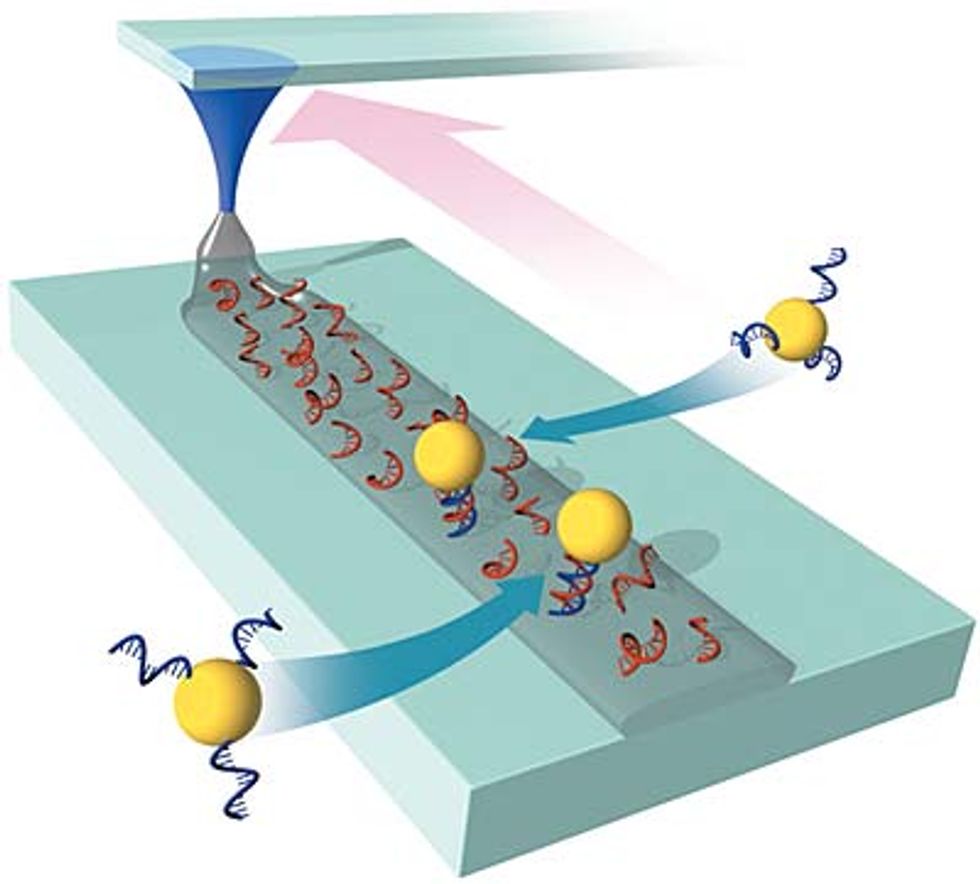
In 2002, Mirkin found the right chemistry to write single-stranded DNA onto glass--a suitable substrate for circuits--with 50-nm precision [see illustration].
Using DPN, he can draw a line of DNA Velcro onto a glass wafer, and a nanoparticle tagged with the complementary sequence will snap into place. "One day you'll literally be able to draw a pattern on a surface, expose that surface to a randomly distributed solution of all the proper building blocks, and they will assemble into some preconceived architecture, perhaps a circuit, a catalyst, a diode, or a sensor," says Mirkin.
Molecular spadework
The road from these early demonstrations to self-assembled circuits with millions of components is a long one. A future circuit might have to self-assemble with the fidelity, for example, to match the gate voltages on each of millions of transistors in a circuit, DuPont's Gierke explains. "Some really major issues have to be overcome," he says.
For her part, Belcher hopes to get there by building even more powerful tools for self-assembly, though via a slightly different route. Because viruses are not really living things--they must hijack the cellular machinery of bacteria to reproduce--their usefulness is limited. Instead, she's shifting to living cells, trying to engineer varieties that can construct and manipulate many types of material. A cell's genes, unlike those of a virus, can encode complex instructions that can be executed in real time. With a microbe like that, engineers could use biology not just to build circuits, but also to diagnose and repair them.
If it all works out, engineers making good on Gordon Moore's famous law--that the number of transistors per IC will double every couple of years--may ultimately owe their success to the biologists Watson and Crick.
To Probe Further
To see the directed evolution process set to music, go to https://neon.cm.utexas.edu/belcher/Research/selection.htm.
The seminal work showing that directed evolution could engineer viruses to interact with semiconductors was "Selection of peptides with semiconductor binding specificity for directed nanocrystal assembly," by S.R. Whaley, et al., Nature, 8 June 2000, Vol. 405, no. 6787, pp. 665-68.