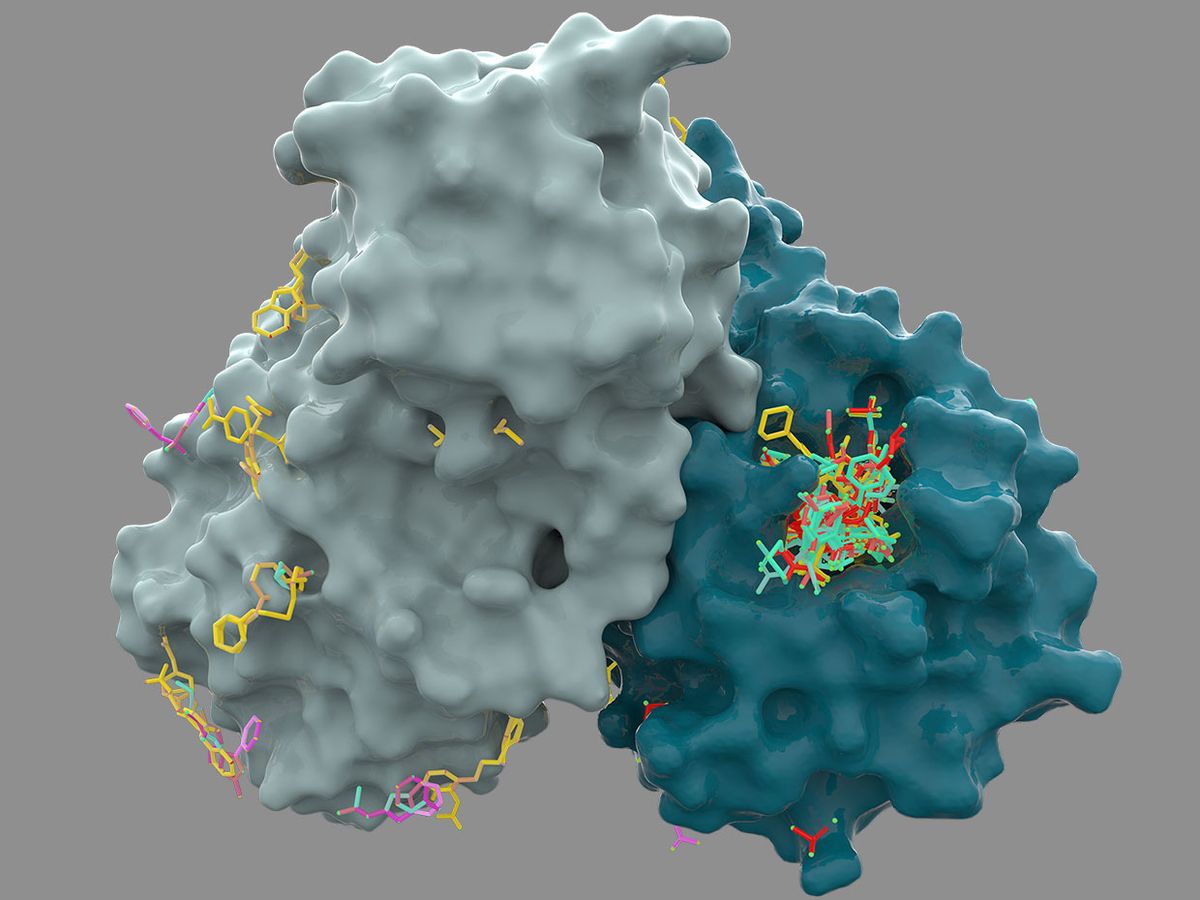
It started with a tweet. Alpha Lee, cofounder and chief scientific officer of machine-learning company PostEra, read on Twitter that Diamond Light Source, the United Kingdom’s national synchrotron facility, had identified a set of chemical fragments that attach to an important coronavirus protein.
Lee wondered if his company, formed just six months earlier, could help connect the dots from fragments to viable drugs to fight COVID-19. PostEra uses AI algorithms to map routes for drug synthesis to speed the drug-discovery process. But to do so, it would need some design ideas. So Lee asked the Internet.
On 17 March, in collaboration with Diamond, the PostEra team launched the COVID Moonshot to crowdsource drug designs from medicinal chemists. Then PostEra applied their technology, pro bono, to determine if and how those designs could be made.
“We thought we might have 100 or 200 submissions,” says Lee, an associate professor at the University of Cambridge. “In fact, we got thousands.”
Over 4,500 molecular designs from 280 contributors around the world flooded into the submissions site PostEra set up for the effort. Two chemical synthesis companies have stepped up to physically make the compounds, providing their services for free or at reduced cost, and two pharmaceutical companies, UCB and Boehringer Ingelheim, are contributing employee time toward the effort at no charge.
“There’s an element of ‘we should do something’ from the PIs of the project, almost a duty, not wanting to leave key scientific equipment and great minds idle, and it has caught on” says John Spencer, a professor of bioorganic chemistry at the University of Sussex, who worked for 10 years as an industry medicinal chemist and is volunteering in the Moonshot. In addition to being involved in discussions and providing advice, Spencer has submitted around 100 ideas and sent compounds from his university laboratory for testing.
If and when any drug candidates are identified via the crowdsourced project, the drug designs will be made openly available in the public domain without patent or any intellectual property restrictions. “In times like these, when the world is closing down, science should open up,” says Lee. “We are really optimistic that we can get a viable [drug] candidate out of this effort.”
Currently, the chemical fragments discovered at Diamond are a far cry from actual drugs. The small molecules only weakly attach to the active site of a key coronavirus protein. A true drug compound requires additional chemical components to be potent, safe, and lasting in the body.
Design ideas for such compounds are being submitted by academics, students, retirees, industry medicinal chemists, and more. “There are a lot of like-minded experts using a diversity of tools: some expert intuition, others machine learning, and others physical modeling,” says Lee.

The PostEra team runs the designs through their machine-learning pipeline—algorithms trained on over 10 million chemical reactions scraped from patents of existing chemicals—to triage which designs can be made, and then generate recipes to do so rapidly. In a 2019 paper, PostEra’s algorithms outperformed human chemists in predicting the outcomes of chemical reactions.
Lee estimates that designing ways to synthesize over 2,000 molecules might take chemists about three weeks. The PostEra algorithms did it in a weekend.

Once the first batch of designs was triaged and complete, PostEra sent them off to chemical synthesis companies Enamine and Sai Life Sciences, which synthesized the compounds at no or reduced cost. “They’ve been super generous with their time,” says Lee. Incurred synthesis and testing costs are being funded through a GoFundMe campaign.
Next, laboratories at the University of Oxford, in England, and Weizmann Institute, in Israel, began testing the compounds against the coronavirus protein, an enzyme called Mpro that is central to the virus’s ability to replicate. So far, there are several promising leads, says Lee. After testing against the protein in a dish, any strong hits will move into being tested against the whole virus, then against viral infection in animals. Lee hopes to identify a preclinical candidate in the next few months.
“Knowing there’s a chance we’ll stumble upon some new discoveries, in an unprecedented manner, with a number of scientists from all countries, all levels—everyone is welcome, everyone has a voice—is invigorating,” says Spencer. “I sincerely hope that this is a sign of things to come.”
If you want to get in on the effort, the Moonshot team continues to welcome design submissions through their website.
Megan is an award-winning freelance journalist based in Boston, Massachusetts, specializing in the life sciences and biotechnology. She was previously a health columnist for the Boston Globe and has contributed to Newsweek, Scientific American, and Nature, among others. She is the co-author of a college biology textbook, “Biology Now,” published by W.W. Norton. Megan received an M.S. from the Graduate Program in Science Writing at the Massachusetts Institute of Technology, a B.A. at Boston College, and worked as an educator at the Museum of Science, Boston.