Probably the most masterful and mysterious act of chemical computation is when a single cell uses its DNA to divide, multiply, and specialize to produce a fully developed organism. In research reported this week in Nature, computer scientists took a small but important step toward harnessing the potential of chemical computation by constructing the first broadly programmable DNA computer.
The system executes a wide variety of 6-bit programs using a set of instructions written in DNA. The researchers used it to perform 21 test programs, though the system is capable of many more. Previous DNA computer schemes were essentially bespoke systems, only capable of solving the single problem they were designed for.
The new system, which is made of just DNA and salt water, is unlikely to find a technological application itself. But it is a step toward developing self-assembling programmable matter, where chemical software automatically directs the construction of materials with complex, programmable nanometer-scale features. Its creators were “trying to understand how to embed computational behaviors within chemistry in order control what chemistry does,” explains Erik Winfree, the professor of computer science and bioengineering who led the research, which was mostly conducted at Caltech.
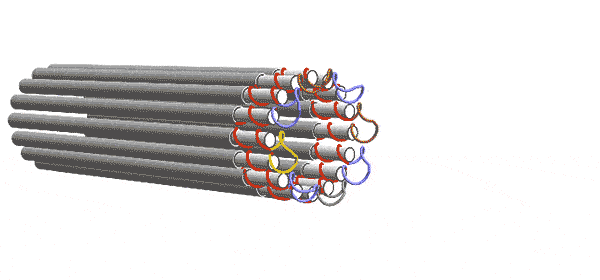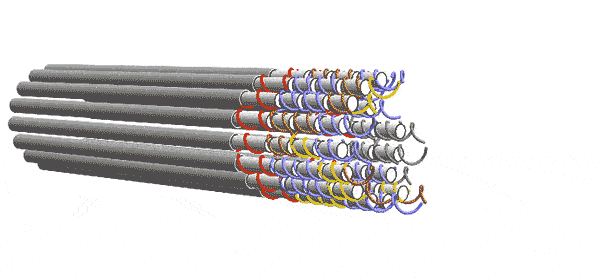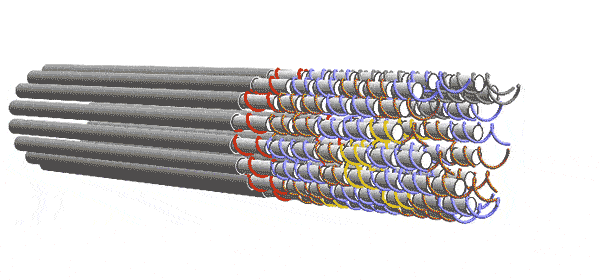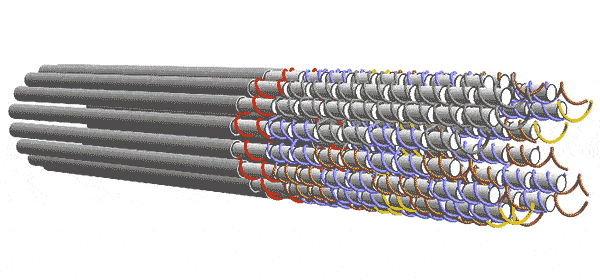
DNA forms a ladder-like structure made up of two long strands. Each strand is a sequence of four chemicals. The computer depends on the fact that single-stranded DNA will try to pair up with other strands having a sequence of chemicals complementary to its own. The Caltech team engineered their DNA sequences to act like 2-input/2-output Boolean logic gates. Five of these gates are connected to form a layer the performs the desired computation with 6 inputs and 6 outputs. But one layer alone can’t complete the computation. Instead, a series of functionally identical layers, with the outputs of one connected to the inputs of the other, perform the computation iteratively until it converges on an answer. In other words, the algorithm progresses as layer after layer links up, essentially knitting a nanoscale scarf that contains every step of the computation.
Winfree and his team conceived of these circuits in the abstract as square tiles with fixtures on their sides that would only allow them to link to a tile with a complementary fixture, as though the tiles were puzzle pieces. The computer’s complete set of instructions comprised 355 different types of tiles. In reality, each tile was made of four single strands of DNA rather than one, thereby adding enough redundancy to overcome many errors.
In order for the computation to progress in an orderly fashion—essentially growing a crystal that computes—the researchers needed something that would act as a starting point and a structure on which the DNA could assemble. “We want the DNA to stick together to do the circuit, not [stick together] randomly in solution” says Damien Woods, who worked on the DNA computer and is now at Maynooth University in Ireland.
The answer was more DNA, explains David Doty who also worked on the computer and is now at University of California Davis. This DNA made use of a materials technology called DNA origami, and it was designed to fold itself into a 150-nm-long, 20-nm-wide nanotube. The components of the nanotube acted as a sort of seed on which the computational crystal could assemble, similar to the way rock candy crystallizes at the end of a sugar-coated string.

Running a program on this 6-bit DNA computer starts with a custom-built compiler. “A huge amount of thought and science went into the compiler,” says Woods. It allows the researchers to write whatever software algorithm they want and translates that algorithm through the various levels of abstraction: logic to tiles, tiles to error-correcting sub-tiles, sub-tiles to DNA sequences, and so on. Finally, it produces what is essentially a recipe of what DNA sequences to add, when to add them, and at what temperature. It takes one or two hours for the DNA origami nanotube to grow and another day or so for the computation to complete. (Speed was never the point, Winfree notes.)
The origami structure was key to reading out the answer. Its DNA sequence was designed so that everywhere in the computation where the bit equals 1, a protein label would attach. The nanotube also unzips to form an easy-to-examine rectangle. Under an atomic force microscope, you can then read the progress of the computation through to the final result presented as protein-labelled bits.
For Winfree, this result has been a big step in a full-career quest. “This has been, for me, quite a long slog,” he says. His interest was sparked as a graduate student about 25 years ago, when he became intrigued between a curious connection between mathematical tiling theory and theoretical computer science, and he wondered if it extended to models describing crystal growth. “It made me intensely curious if pseudo-crystals carry out computations as they grow.” Now we know that they can.
Samuel K. Moore is the senior editor at IEEE Spectrum in charge of semiconductors coverage. An IEEE member, he has a bachelor's degree in biomedical engineering from Brown University and a master's degree in journalism from New York University.