When you come down to it, designing a lab-on-a-chip (LoC) strongly resembles managing a full-scale biomedical lab: Samples must be separated, technicians assigned, instruments scheduled, and apparatus cleaned. All of this has to be optimized to keep it running at peak efficiency, even when techs are out sick, machines go AWOL, and uncleaned glassware stacks up.
Michigan Technical University (MTU) researchers Chen Liao and Shiyan Hu are members of the relatively small group of researchers meeting the biochip-optimization challenge by developing computer-aided design (CAD) tools. (A Google Scholar search for “CAD biochip” turns up 243 papers published since the beginning of 2013.) Specifically, the MTU engineers are building software to improve the physical layout of the "discrete-droplet" lab-on-a-chip. They reported their latest work in IEEE Transactions on Nanobioscience.
There are two basic kinds of biochips: Continuous flow chips have “permanently etched micropumps, microvalves, and microchannels.” And discrete-droplet chips have a two-dimensional array of chambers connected by channels, through which individual droplets are moved via electric-field-induced fluid flow, shunted from box to box by varying charges on electrodes that sandwich the chip. The discrete-droplet design is highly flexible: many droplets can be guided through the chip, like city buses traveling many routes through city streets.
Droplets can be guided into other droplets so their contents can react, then nudged toward yet another cell where a sensor stands ready to measure the results. If properly designed, a single discrete-droplet chip can conduct many different syntheses and analyses almost simultaneously by carefully shuffling droplets with different compositions around one another. Imagine doing a full blood-chemistry panel with a single drop on a single chip.
“In a very short time, you could test for many conditions,” Hu says. “This really would be an entire lab on a chip.”
But that “careful shuffling” is the rub: collision, contamination, variation, and dead-ends lurk at every corner, confounding simple routing and scheduling. For example, droplets can never be in adjoining boxes. No droplet should occupy a box or run down a channel until the last vestiges of the previous droplet have dissipated. Reactions can run faster or slower with changes in temperature or humidity. Some measurements can take longer than others. And fabrication errors or later damage can put some boxes and channels out of commission entirely.
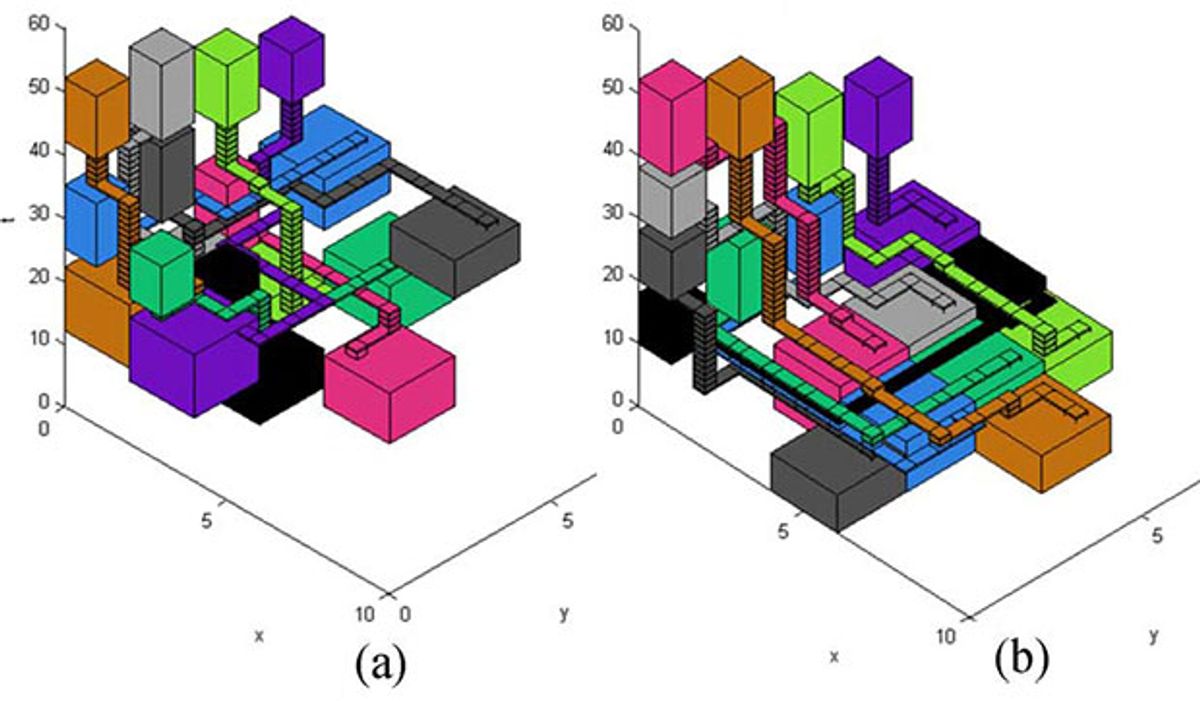
Liao and Hu map the chip’s surface, with its wells and channels, onto the x-y plane. Then they add a vertical axis of time. Every process then becomes a path through the space-time cube. Constraints mean that paths can never intersect. To prevent cross-contamination, a cocoon of open space and time must surround every path. The buffer around variable operations—such as DNA amplification or protein synthesis—increases still more to accommodate changes in timing. And some paths are blocked off completely to avoid blocked channels and damaged boxes.
Though earlier research had also constructed “contamination aware and defect tolerant” biochip algorithms, Liao and Hu say theirs is first to consider the effects of process-time variation.
To test the concept, they optimized designs, both with and without considering the effects of operation variation, and then ran a series of simulations to assess how each approach performed. The measuring stick was “routing yield,” the percentage of test runs that go successfully from beginning to end as process conditions varied. While the variation-tolerant process paths were slightly longer, the difference in success rates was striking. Designs that ignored process-time variation succeeded 15 to 62 percent of the time. Variation-aware processes scored 100 percent. Overall, an increase in process time of 3.5 percent produces a 51 percent jump in total throughput.
“It has taken us four years to do the software, but to manufacture the [lab-on-a-chip] would be inexpensive,” Hu says. “The materials are very cheap, and the results are more accurate than a conventional lab’s.”
Douglas McCormick is a freelance science writer and recovering entrepreneur. He has been chief editor of Nature Biotechnology, Pharmaceutical Technology, and Biotechniques.