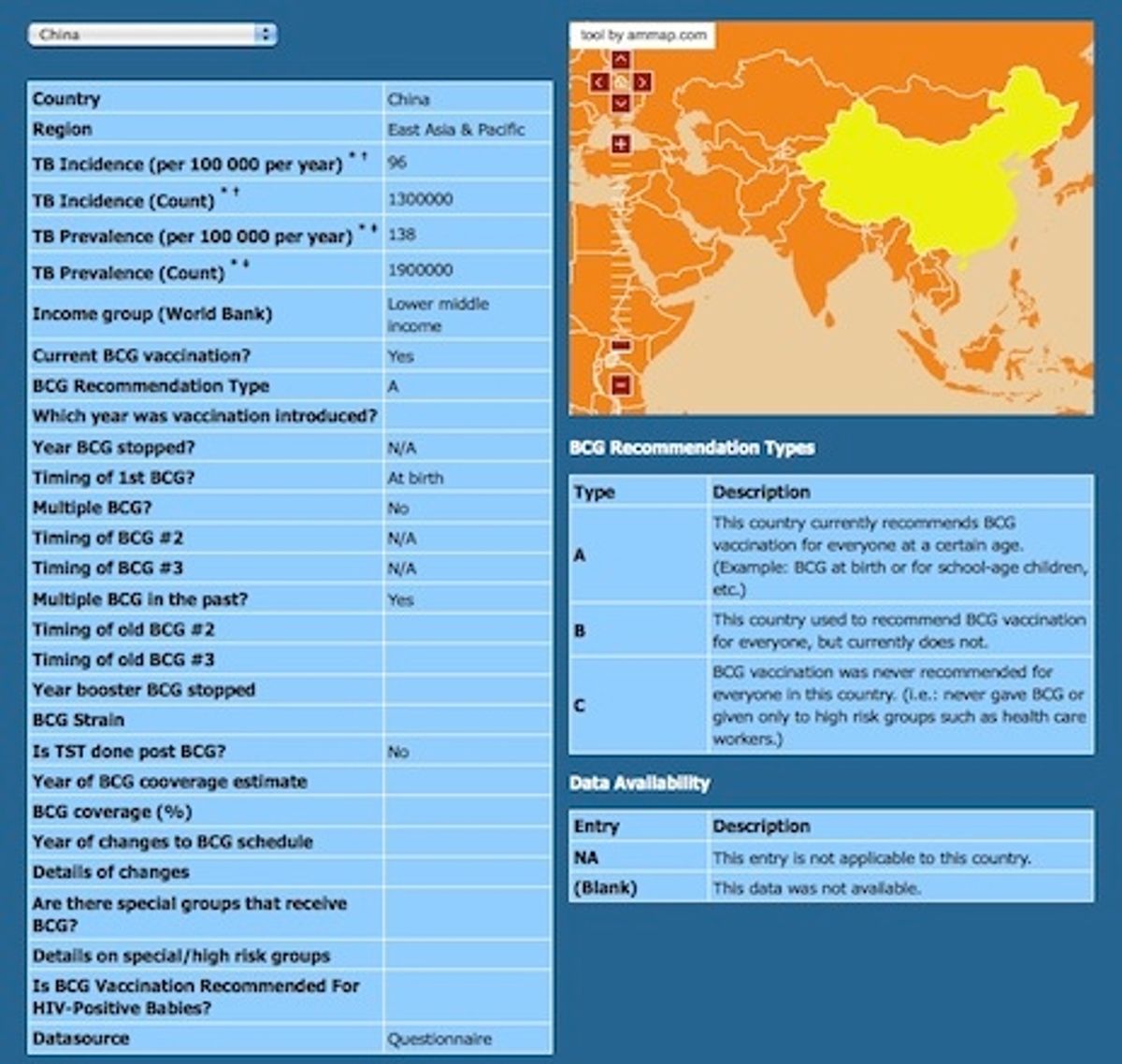
Tuberculosis is notoriously difficult to diagnose, so researchers are always looking for ways to make the detection process more efficient. One problem that clinicians have struggled with is false positives on the commonly-used TB skin test that can occur in people who were vaccinated against TB as children.
Not all countries use the TB vaccine--the US and Canada, for example, have never used it nationwide--but plenty of others do, and patients often don't remember which vaccines they've had. Now researchers at McGill University in Montreal have created an interactive map that merges disease statistics with TB vaccine policies from 180 countries. The goal is to help ease clinicians' confusion over whether a patient might have been exposed to the vaccine.
Interactive maps have become a popular way to display data in the infectious disease field. The World Health Organization has developed several maps, including one that international travelers can use to determine malaria and rabies risk, and Google Flu Trends helped clinicians and patients keep track of the H1N1 epidemic in 2009. But few have taken the extra step of incorporating policy data.
In the case of the TB vaccine map, adding the policy data was essential because there were no other databases clinicians could turn to for that information. Indeed, the McGill researchers wound up collecting most of the policy data first-hand through surveys and literature searches.
The next challenge for the McGill team will be to enhance the map with country-specific data on other diseases--mainly HIV--that affect TB diagnostics and response to the TB vaccine. The project is described in detail in an article published in PLoS Medicine last month.