GM Foods Grow Up
With help from robots and new genetic tricks, farmers could feed the planet while dodging the controversy

The stalks of foxtail millet were bent under Guangdong province’s hot summer sun. The plants were heavy with seed, giving them impressively bushy “tails” that would have done any fox proud.
Archaeologists believe that people began cultivating foxtail millet in China as early as 6500 B.C.E., but the plant looked considerably different in those Neolithic days—the foxes’ tails were thin and scrawny. Nevertheless, this early cereal crop had many things going for it, and researchers believe the hardy, quick-growing millet was more common than rice in China’s arid north for millennia. But rice, with its high yield of grain, gradually won out, and the Chinese nearly forgot all about millet—until now.
Researchers at the Chinese genetics company BGI, which planted the bushy millet in Guangdong, say this “neglected crop” could help their nation meet its modern agricultural challenges. That’s why they recently began a breeding program to augment this plant’s useful traits while also boosting yields. “It’s very drought tolerant,” explains Zhang Gengyun, general manager of BGI’s life-science division. “So I think that this plant could be valuable in the future, especially with conditions of global climate change.” He adds that little scientific work has been done on foxtail millet thus far, “but thanks to today’s genetic technologies, we can explore the potential of the species.”
No company is better positioned for that exploration than BGI, which has leveraged the Chinese government’s financial support to grow into one of the world’s biggest genomics companies in less than 15 years. BGI specializes in sequencing whole genomes—the entire complement of genetic material that makes up the DNA of a species. Its research facilities are packed with dozens of high-tech sequencing machines, and more than 800 employees work to analyze the resulting data with the latest bioinformatics software. BGI collaborates with institutions around the world, bringing its sequencing firepower to bear on the genomes of microbes, plants, animals, and humans.
BGI is just one of many companies interested in plumbing the genomes of the world’s crops to launch another green revolution, this one enabled by a level of precision engineering and automation that agricultural research has never enjoyed before. As institutions like BGI create a deluge of genetic data, seed companies are using that information to quickly design new supercrops. Together they’re changing assumptions about how much food farmers can grow and where they can grow it.
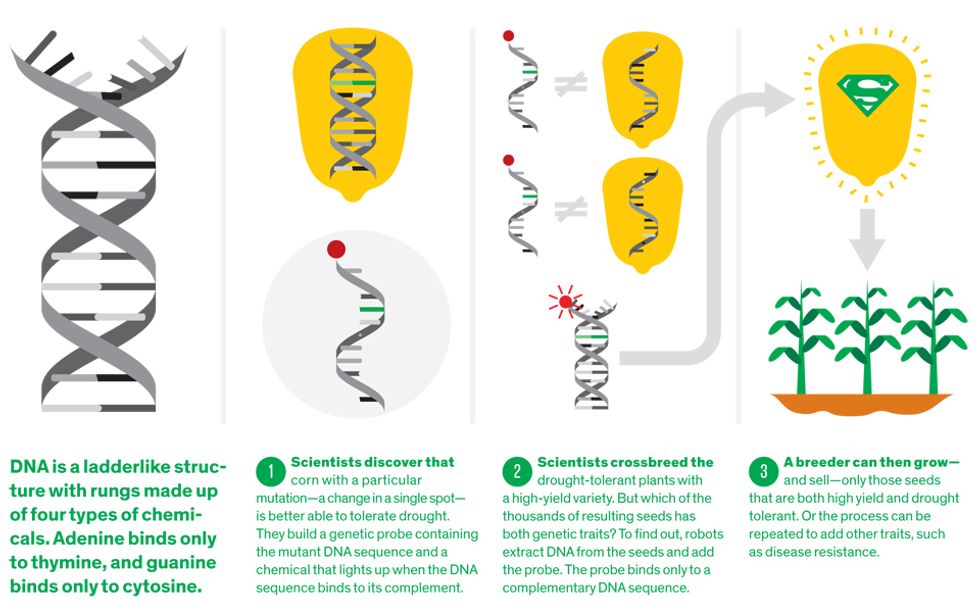
It all starts with the same material. Whether researchers are studying millet, mouse, or man, the genetic instructions they seek to decipher are coded in a long molecule called DNA. The code consists of a sequence of chemical bases, which come in four flavors: adenine, thymine, cytosine, and guanine, abbreviated as A, T, C, and G. The whole long series of letters is known as the genome. Specific pieces of the genome—certain strings of letters—are the genes, which carry the important instructions about how to make proteins.
Genome sequencing is the process of decoding these vast sequences of letters. In agricultural research, that information helps botanists add certain desirable traits to a plant by carefully controlling its genetics, either by traditional crossbreeding methods or by inserting genes directly into the genome. When researchers use the latter method to move genes around, the resultant “transgenic” crop is considered a genetically modified (GM) organism, and controversy ensues.
Critics of GM foods have questioned their effect on human health, the environment, and the economic well-being of farmers. The naysayers worry primarily about crops that include genes taken from another species entirely, saying that scientists can’t predict all the consequences of such meddling. Activists have used guerrilla tactics to make their displeasure known, sometimes making midnight raids on field trials of GM crops to uproot and trample the “Frankenfood” plants. Even the most humanitarian efforts to increase crops’ nutritional benefits have met with impassioned resistance [see sidebar below, "Golden Grains of Discord"]. But as the tools of genetic engineering get more sophisticated, it’s becoming less clear how to define what a GM crop is and whether it requires scrutiny and regulation. In this article we’ll describe how computers and robots are advancing those tools and leave questions about the appropriate level of regulation to others.
The threat of provoking controversy has kept BGI from experimenting with the types of GM crops most likely to raise objections—those that mix genes between species. The company’s researchers sometimes use transgenic techniques to take a gene of interest from one plant and move it to another of the same species, which allows them to check whether the gene really causes the effect they think it does. But Zhang says the company is primarily interested in using genomic tools to speed up traditional breeding methods. “We can tap the potential that already exists in the genome,” he says.
The foxtail-millet project, Zhang explains, was intended to demonstrate the potential of BGI’s sequencing technology to revolutionize agriculture. Zhang’s team spent about four and a half months and US $1.5 million to sequence the plant’s genome and published the results in Nature Biotechnology. The BGI team took another two months to complete genetic maps showing which stretches of sequence controlled which traits in the plant, then crossbred plants to create seeds with the exact mix of traits they sought. One and a half years after they began sequencing the millet genome, BGI researchers were admiring their hearty, high-yield crops in that research field in Guangdong province.
BGI has already begun to apply the lessons learned from that project to a much larger endeavor. In cooperation with the International Rice Research Institute (IRRI), based in the Philippines, the company has embarked on an ambitious effort to sequence the genomes of 10 000 strains of rice. The researchers have already produced the first 3000 genomes and are now working to link that genetic information to the specific traits of each strain, a process known as genotyping. Researchers and breeders can then create crops that are customized to produce high yields while meeting specific local challenges, like saltwater flooding, drought, or particular pests.
Eero Nissilä, head of IRRI’s Plant Breeding, Genetics, and Biotechnology Division, says the rice project is critically important to the future of humanity. “What worries us is that we expect 9 [billion] or 10 billion people to be living on this planet by 2100, and half of them eat rice as a staple food,” says Nissilä. “We think there’s no way to feed them all if we don’t increase the yields of rice.” But, he says, researchers can dive deep into the diverse gene pool of rice—there are 24 species and up to 500 000 varieties within those species—to find plenty of useful traits. Until recently “this research would have been impossible,” Nissilä says, “but with today’s technology it’s not even that expensive.”
“Twenty years ago, if I was to try to put 20 traits together in the same plant, they would have laughed,” says Robert Fraley, chief technology officer at the world’s biggest biotech seed company, Monsanto. “The odds of that working would be one in a trillion.”
No one’s laughing now. Monsanto, with some assistance from competitor Dow AgroSciences, engineered resistance [PDF] to corn rootworm, defense against caterpillars, and tolerance to two kinds of herbicides into maize seed it released in 2009. Switzerland’s Syngenta discovered varieties of 13 genes that enhance maize’s ability to withstand drought and, using its proprietary cycle of breeding and genotyping, inserted combinations of them into high-yield breeds that were put through their paces during a drought that parched much of the United States in 2012.
What’s changed in those 20 years? A lot, of course—new knowledge of plant physiology, made-to-order DNA sequences, and a host of other biomolecular advances. But it’s what’s happened in the past five or so years that’s shaping the future. And that can be summed up in one word: robots.
If you want to find what’s probably the best example of what robots are doing in crop development, come to the epicenter of American agriculture—Iowa. That’s where DuPont Pioneer opened a new $40 million production genotyping lab last year.
The lab, like many things this far out from Des Moines, is surrounded by fields of rich-looking black topsoil. That topsoil, along with Iowa’s climate, are “why we’re all here,” says a lifelong Iowa resident. On the steel-gray October morning that IEEE Spectrum visited, the beige stubble of last season’s experimental maize was still visible in the fields outside. Inside the lab, several clusters of whirring, chugging, and occasionally banging machines do their work.
Bot-Assisted Breeding: IEEE Spectrum tours Dupont Pioneer's production genotyping lab to see how the process of improving the genetics of crops is automated.
What goes on here is akin to what happens in one of the server farms that run Google’s search engine. The lab is an information factory whose primary purpose is to answer a single question more than a 100 million times per year: Does this seed contain variants of genes that are associated with these traits? The difference is that the database is, quite literally, the DNA of various crop strains and the search is executed using biochemistry. Armed with the answers to their queries, breeders can know whether it’s worth growing that seed and which of thousands of other seeds to cross it with.
The basic procedure is this: First, DNA is extracted from a bit of a plant and unzipped down the middle, so that a complementary sequence can bind to it. (In the search engine analogy, that DNA is the database.) Then short sequences of DNA, called genetic markers, are added. (These are analogous to the search terms.) Usually not the complete genes themselves, these markers code for distinct patches of DNA in the plant’s genome that are of interest to breeders. For instance, a particular sequence might be found in all maize that can survive in near-desert conditions but not in any that can’t. The marker, which is chemically labeled so it can be identified later, will stick to the plant’s unzipped DNA if it’s a match or wash away if it isn’t.
Decades ago, it was a tricky procedure. Now, what was once months of work that would earn you an advanced degree is done tens of thousands of times each day. At Pioneer’s lab, robotic arms swing to a hypnotic beat, moving cassettes of plant material from one step to another, adding just the right amount of just the right chemicals and markers to each of thousands of DNA samples. All the while technicians and scientists keep track of the results on giant screens.
100 million
Number of genetic experiments DuPont Pioneer’s new roboticized genotyping lab can perform per year
6 terabytes
Maximum total daily data output of BGI’s sequencing machines
US $500 000
Cost of each of 128 sequencing machines BGI purchased in 2010
What’s all this automation good for? The most easily measurable metric is that “running a query” now costs mere pennies instead of hundreds of dollars. “We couldn’t do hundreds of millions of data points a year if it cost what it did five years ago,” says John Arbuckle, senior research director for Pioneer’s production genotyping facilities. That means more varieties can be screened and breeders can make a better selection of what plants to cross-pollinate with which. “Out of the thousands of possible plants to put out into the field, we can choose the fraction that is likely to succeed,” he says. That means getting not just a better crop—“increasing the rate of genetic gain,” he calls it—but also a faster one. “What used to take seven or eight generations to get now takes three to five.”
To fully appreciate the change, you have to understand what an insanely frustrating experience breeding was before industrial-scale genotyping. If you wanted to see if any of your soybean strains were vulnerable to a particular disease, you couldn’t just query their genomes to see if any had the protective genes. You had to find a farm where the disease had occurred in the previous season, rent out the field, and “then pray that the weather would be right for that disease to occur,” says John Soper, Pioneer’s vice president of crop genetics R&D, who started his career as a soybean breeder in the 1980s. “And 9 times out of 10 it didn’t.”
On the economic side, the technology can be used to improve the yield of crops for markets that were heretofore too small to catch the attention of the big seed firms. It makes “the technology more accessible to the crops and regions that in the past haven’t been able to afford it,” says Arbuckle. Those markets might be characterized by a peculiar mix of conditions—soil, weather, pests, temperature—that don’t occur elsewhere, and can’t be served particularly well by what companies already have on hand. For instance, Pioneer is part of an effort led by the United States Agency for International Development to come up with maize that can thrive in the million hectares of South Asia that experience excruciating heat during the crop’s crucial flowering season. Genotyping can enable a breeder to predict which maize has genes that might make it flourish in such conditions and breed those into a high-yield strain.
Often you can get everything you want from a plant’s own genome, but sometimes you just can’t. There are categories of problems for which no good solutions exist in a plant’s genome. Insect resistance is generally such a problem, says Soper. Or if you want to add a nutrient to a crop that just doesn’t make it, you’re going to have to add some new blueprints to that crop’s genome.
The tried-and-true method of genetic modification involves a particularly useful pest called Agrobacterium tumefaciens (or Rhizobium radiobacter if you’re feeling formal), a soil bacterium that causes tumors in plants. How it forms those tumors is the interesting part: It injects a small segment of DNA into the infected plant’s cells, and that DNA gets incorporated into the cell’s genome. Starting in the late 1970s, scientists realized that they could engineer the agrobacterium’s delivery system to inject useful genes such as the one for the toxin made by the microbe Bacillus thuringiensis (thus insect-resistant corn and cotton) or for an enzyme unaffected by the herbicide glyphosate (thus Roundup Ready soybeans).
It’s not quite that simple, of course, and the method has some hang-ups. The agrobacteria inject their DNA payloads into individual cells, so to get a whole genetically modified plant, you often have to grow it from a cell culture. This isn’t just time-consuming and inefficient—there are many plants that scientists don’t know how to grow from cultures. (Hence, no genetically modified peppers.) Equally irritating is that agrobacteria are not too particular about where their injected DNA goes in a plant’s genome. The injected DNA gets incorporated in semirandom spots, meaning it can land in a place where the cell’s machinery might never put it to work. Or worse, it might scrunch itself right into the middle of a gene that the plant actually needs to survive.
But in the past two decades, agricultural biotechnologists have come up with ways of manipulating a plant’s genome much more precisely. One way puts to work a class of natural nanomachines collectively called restriction endonucleases. An endonuclease is effectively a smart pair of molecular scissors. It finds a particular sequence in a cell’s genome and then snips.
When a double strand of DNA is cut, the cell’s molecular machinery tries to repair the damage. Snip the DNA in two places and the cell might join it together without the snipped-out part, disabling or at least mutating a gene. Do the same in the presence of a new DNA sequence—say, one that makes baby’s breath flowers mauve—and the molecular-repair machinery might splice it right into the desired spot. “The cell knows how to put together broken pieces,” explains Alexander Vainstein, a plant biotechnologist at the Hebrew University of Jerusalem. “But it doesn’t know what the original pieces were.”
In the last few years, made-to-order endonucleases have gone on the market, and biotechnologists have started using them to great effect. Dow AgroSciences, for example, used a type called a zinc-finger nuclease to delete part of a maize gene. The end result was to reduce the maize’s level of phytate, a so-called antinutrient because it makes it harder for animals to absorb critical minerals.
Vainstein has a solution to the other problem with the earlier generation of genetic transformation: having to grow the modified plant from a cell culture. His lab and the Israeli biotech company Danziger Innovations have invented a strain of virus that infects whole leaves or even an entire plant with a temporary genetic blueprint for an endonuclease. So for a short period, the cell’s own machinery builds the enzyme, which does its gene-editing job. The virus burns itself out, and the infected plant’s seeds carry the genetic modification forward. (The first genetically modified pepper is now in the works at Vainstein’s lab.)
A side effect of the new, more-precise genome-editing technology is that it’s raising questions about how genetically modified crops should be regulated by governments. For instance, tolerance to the herbicide glyphosate has so far been engineered into commercial crops by transferring in a gene from a bacterium. But some plants, as a growing glyphosate-tolerance problem shows, already carry a much less potent variant of that gene, explains Jennifer Kuzma, an associate professor in the Center for Science, Technology, and Public Policy at the University of Minnesota, Twin Cities. If scientists were to edit the plant’s own gene using the new molecular methods so that it functioned more like the bacterium’s gene, would the resulting resistant plant really be GM?
Here’s one that’s even more of a head scratcher: Plants that get their traits via mutation by chemicals or radiation have been around for more than half a century and haven’t been regulated as GM. But endonuclease methods allow scientists to target a mutation—a change in a single letter of the genome, say—to a precise location in the chromosome. Should the result be considered a GM plant or just the equivalent of a really lucky mutation? “There isn’t a bright line,” says Kuzma. But, she says, “I’m not sure the public will make a distinction” between the old GM and the new. Already, the U.S. Department of Agriculture has quietly decided not to regulate [PDF] at least one experimental crop in part because scientists used one of the new precision methods in its creation.
The line between traditional breeding and genetic modification is indeed getting very blurry—which is not to say that genetic modification is going away. Many farmers and technologists don’t see a way to feed an inevitably more populous planet without it. Some even see it as a necessity for a future after oil extraction becomes uneconomical. Marc Van Montagu, who codiscovered the agrobacteria transformation method, is convinced that “plants will be the raw materials for our chemical industry.” Syngenta has already developed a breed of maize engineered to make the amylase enzyme, an additive that helps break down corn sugars in corn-to-biofuel factories.
“Twenty-first-century plants,” says Van Montagu, “will be GM plants.”
This article originally appeared in print as “An Industrial Revolution in Genetics