Understanding the Human Genome
Rough drafts of the human genome were developed some two years ahead of schedule, but the hard work of understanding what it means is only beginning
At least since Alan Turing tackled Enigma in World War II, building machines to crack codes has been the domain of computer scientists and engineers. Lately they have joined biologists in cracking humanity's most important code--the human genome, the complete set of all our genetic information.
Sequencing the human genome is essentially putting in order the over 3 billion chemical units that encode the instructions on how to build and operate a human being. But those instructions are written in a language biology does not fully understand. Indeed, some have described the genome as a parts list minus information on how the parts connect or what they do. And leading scientists are quick to point out that just knowing the raw data set that makes up the genome is not an end in itself.
Rather, the usefulness of the genome will emerge only after scientists have figured out how the parts go about making the machine that is the human body. This is what the biology of the new millennium is all about. It is an accelerated science based as much on bits and semiconductor chips as on microscope slides and test tubes. "The in silico approach will eclipse in vitro and even in vivo," Francis Collins, director of the National Human Genome Research Institute in Bethesda, Md., predicted. Scientists suspecting a genetic correlation with disease can now seek out starting points in the genes of humans and other creatures, compressing what would have been a decade or more of research into a day or two of database queries.
In fact, an industry known as bioinformatics has grown up around the idea that biology will increasingly depend on sorting and manipulating huge amounts of data. Industry analysts forecast that the market for genomics information and the technology to use it will reach an annual US $2 billion by 2005.
The genetic code cracking, or sequencing, is being done in the main by two groups of scientists: the Human Genome Project and Celera Genomics of Rockville, Md. The Human Genome Project, an international public consortium with funding over 15 years of about US $3 billion, downloads all its sequencing data every day to freely accessible databases run by the National Center for Biotechnology Information, the European Bioinformatics Institute, and DNA Data Bank of Japan. Meanwhile, Celera plans to sell its genome database and related information to pharmaceutical and biotechnology firms.
Both groups announced the equivalent of rough drafts of the entire human genome last June. The Human Genome Project expects that it will take until 2003 to produce a fully polished version, but Celera plans to finish up in 2001.
Celera's and the Human Genome Project's efforts to sequence the genome have often been described as a race--a metaphor that irks most scientists in the public project. Harold Varmus, president of Memorial Sloan-Kettering Cancer Center in New York City and former director of the National Institutes of Health, in Washington, D.C., instead suggested a historical metaphor. "It's a little bit like a new continent," he told reporters in New York City last spring. "The human genome [is] being explored by the Dutch East India Company on one hand and the U.S. Geological Survey on the other."
DNA Basics
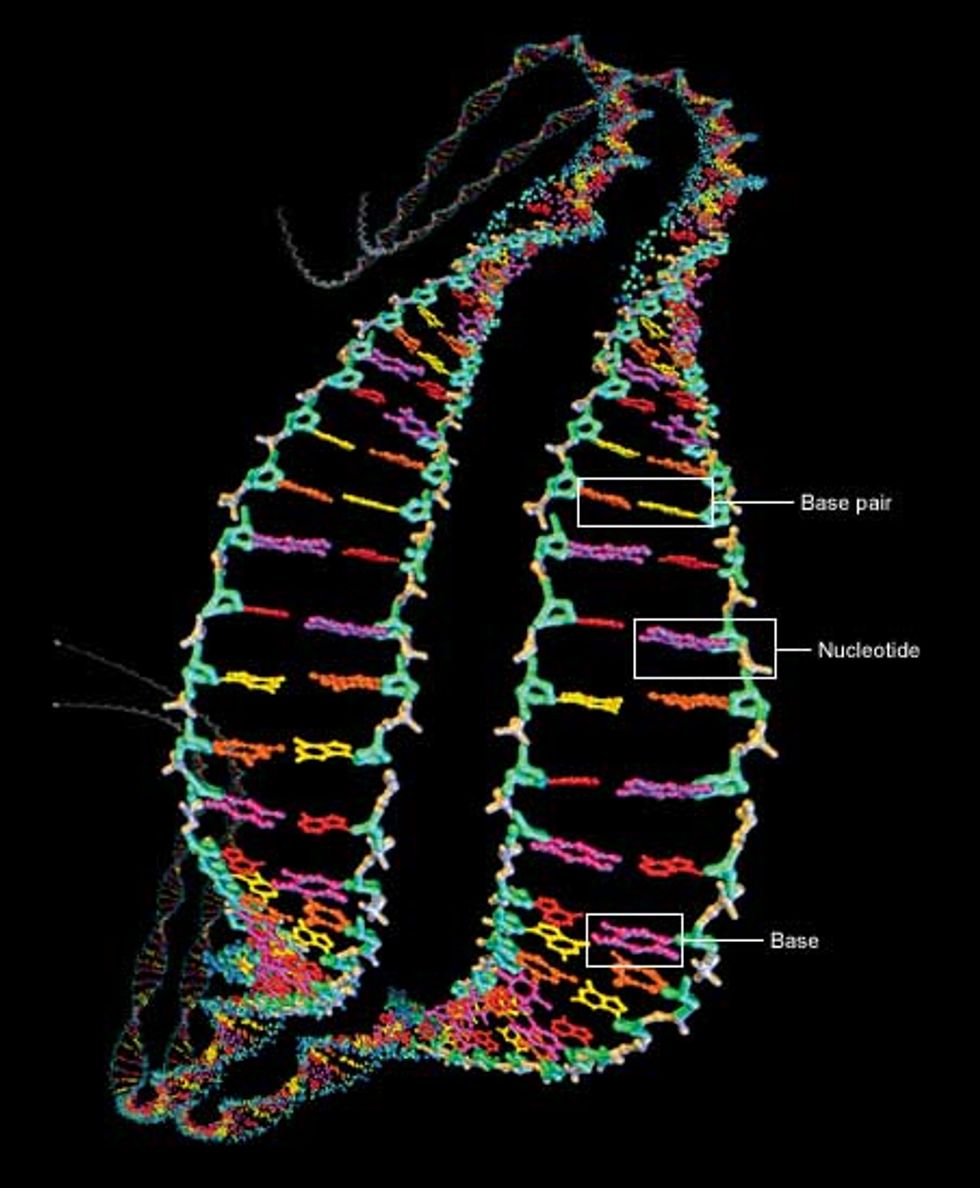
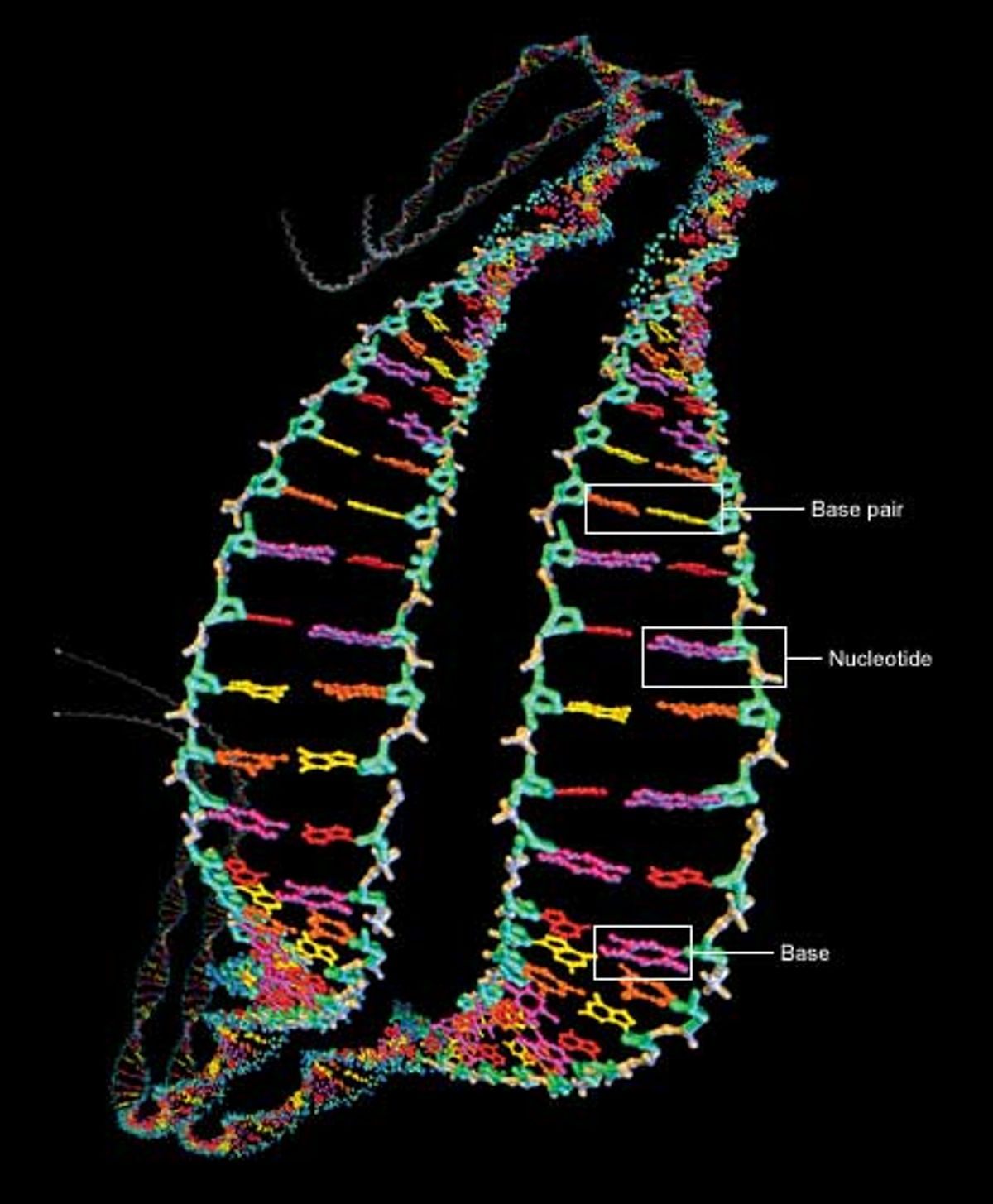
At the heart of the genome sequencing effort is the macromolecule responsible for carrying hereditary information: deoxyribonucleic acid, or DNA. It contains four key chemicals, adenine, thymine, guanine, and cytosine. Referred to as A, T, G, and C respectively, they are each attached to a molecule of deoxyribose sugar plus another molecule of phosphate to form a unit called a nucleotide [Fig. 1].
The nucleotides are the backbone of DNA. They connect up into dual chains that wrap around each other to form a twisted ladder, or double helix. The rungs of the ladder are formed by the A, T, G, or C units of the nucleotides, which are called bases. The bases bind to each other, adenine needing to be across from thymine and cytosine across from guanine. The A-T and G-C units are known as base pairs. In almost all human cells (sperm, eggs, and red blood cells are notable exceptions), most DNA is found as two versions of 23 contiguous segments, called chromosomes.
Not all the sequences in the DNA have obvious functions. Only certain sequences of bases, called genes, spell out the instructions for making proteins and controlling their production. Proteins also are essentially chain-like macromolecules but they fold into shapes that serve some function in the body. For instance, proteins act as structural elements in body tissue; as gates controlling the flow of current into and out of brain cells; and as chemical switches to control cell processes.
Much of scientists' ability to sequence the genome grew out of knowledge of DNA's structure and chemical nature. When heated, the two strands of DNA's helix separate to form single-stranded DNA. The attraction of adenine for thymine and guanine for cytosine directs the synthesis of strands complementary to each of the separated strands to produce identical double helices. Cells use this scheme to copy all their genes before dividing so that a complete set of genes ends up in each cell. They also use it to produce temporary copies of genes for the production of proteins.
The same scheme is also at the heart of DNA sequencing. It allows a procedure called the polymerase chain reaction (PCR) to rapidly copy strands of DNA.
Not only can complete copies be made rapidly, but copies varying in length by just one base can also be generated. These varied lengths of DNA, which are tagged and identified with a fluorescent dye, become the input to DNA sequencing machines, which do the job of finding the same sequence of bases in the original strip of DNA. Thanks to the efforts of hundreds of sequencing machines at work in laboratories around the globe, the rough drafts of the human genome were developed last spring, some two years ahead of a schedule first announced in 1990.
Ongoing efforts
Annotating the genome by attaching function and meaning to its genes is the next step in the discovery process. But first the genes must be located, separating them from the 95 percent and more of the genome that appears uninvolved in the workings of human cells. There are an estimated 25 000-150 000 genes spread throughout the billions of bases that make up the genome, and little is known about the majority of them.
The rough drafts of the human genome were developed last spring, some two years ahead of schedule.
An important assist to the annotation process has been the sequencing of the genomes of other species. Owing to decades of study, many genes and their functions have already been identified in fruit flies and mice, and estimates of the similarity between the genes of mice and men run high. So by matching up known genes in mice or flies with sequences in the human genome, scientists get a clue to the function of human genes. Celera recently worked on the fruit fly commonly used in genetics experiments and sequenced the 120-million base pairs of its genome as a test, before tackling the human genome. Both that company and a public-private group, the Mouse Sequencing Consortium, have now moved on to mice, which have about the same number of base pairs--about 3 billion--in their genome as humans. Celera announced that it completed a rough draft of the mouse genome last month.
Measuring the genetic variation amongst humans is also fast becoming a key outcome of sequencing the genome. That variation in the sequence of bases is thought to be very slight; perhaps only 1 base in 1000 will differ from person to person, making us 99.9 percent identical.
The places in the genome where a gene is spelled differently from person to person are to be cataloged by a consortium of drug companies, computer firms, and public institutions. These genetic locations, called single nucleotide polymorphisms (SNPs), could be the keys to why some people are prone or resistant to certain diseases. At last count the consortium identified 296 990 SNPs. Celera, whose business depends on such value-added information as SNPs, claims to have found at least 2.4 million more.
Biologists, computer scientists, and engineers have a great deal of work ahead of them to make use of the raw data coming out of the project daily. "This is a very rich and complicated text that has had glosses on its meaning added" over the 3.5 billion years since life began on earth, Eric Lander, director of the Whitehead Institute in Cambridge, Mass., one of the main sequencing centers, told reporters last spring. "And it's very unlikely that we are going to be able to extract its full meaning in the course of the next month, year, or frankly, century."
In the article that follows, contributing editor John Hodgson examines the technological innovations that allowed scientists to sequence the human genome. Hodgson explains the workings and development of the machines that performed the sequencing, the automation involved in handling and preparing genetic material, and the computer programs that are assembling the fragmented data sets into a more or less complete genome.