Synthetic Biology’s Hunt for the Genetic Transistor
How genetic circuits will unlock the true potential of bioengineering

In 1977, a small group of researchers in California changed the world when they wrangled a common gut bacterium into producing a human protein. Using every technique in the book—and inventing some of their own—they scavenged, snipped, and glued together genetic components to synthesize a tiny filament of DNA. They then inserted the new segment into some Escherichia coli cells, tricking them into making the human hormone somatostatin.
A year later, these scientists had an E. coli strain that produced insulin, an invaluable drug in the treatment of diabetes. With that, the era of biotechnology was born. A plethora of novel—or at least cheaper—drugs seemed to loom on the horizon.
Thirty-odd years on, molecular biologists have delivered on many parts of that early promise, engineering microbes to produce a wide range of pharmaceuticals, including experimental antimalarial medicines and antibiotics. A quick glance in the pantry or storage closet is likely to reveal other products of genetic engineering, too, including foods, food additives and colorings, and even laundry detergent. The list goes on and on.
The economic impact of all this has been enormous. Genetic engineering and other forms of biotechnology account for some 40 percent of the recent growth in the U.S. gross domestic product, for example. The biotech sectors in other countries have also made sizable contributions to their economies. And you can expect that trend to continue as genetically engineered organisms tackle even more diverse challenges, such as producing renewable fuels and cleaning up toxic waste.
Genetic engineers have indeed accomplished a great deal, but they’ve also run up against many obstacles in transforming microbial cells into factories that churn out useful substances. In a real-world factory, you need all your production machinery and employees operating in sync to run an efficient business. A cell also has components that act like machines, producing complex biological molecules, and other parts that act like messengers, ferrying around information in the form of chemical signals. Bioengineers have to do quite a bit of tricky fine-tuning to their cellular factories, manipulating the operation of many subcellular components so the cells don’t die as they crank out the desired product in copious amounts.
For the bioengineers who do that tuning, a more apt analogy than the factory is that of an electronic circuit. We essentially want to make a cell programmable. We’d like to give it a command and have it perform a new function, just as if it were a tiny computer. To gain this power, we first need to amass a collection of well-characterized biological circuit elements that we can arrange however we like. This is one core focus of a subset of bioengineers, known as synthetic biologists, who are trying to revamp how genetic engineering is done.
Synthetic biologists take their circuit analogy quite seriously, despite the fact that a cell is a whole lot squishier than a silicon wafer or circuit board. A cell is basically a little bag full of biomolecular signals that cause cellular machinery to read chunks of DNA and with that information produce other useful biomolecules, typically proteins. Millions of different kinds of proteins are found in nature, and they participate in countless cellular processes, including the very ones that govern their production.
In a cell, certain proteins can influence pieces of DNA that code for something else. Those gene products can in turn affect other stretches of DNA and so on, forming complex webs of biochemical interaction. Think of these as the genetic circuits making up the cell’s CPU. If you want to program a cell to do something specific, you need a way to build—or at least control—these genetic circuits.
Developing the requisite cellular control mechanisms is one of this century’s great technical challenges. The two of us belong to two of the largest academic groups pursuing this goal: the Synthetic Biology Engineering Research Center, based at the University of California, Berkeley, and the BIOFAB: International Open Facility Advancing Biotechnology, which has its headquarters in nearby Emeryville, Calif. The goal of both projects is to help genetic engineers configure organisms the same way electrical engineers configure complex circuits. You could say we’re trying to put some honest-to-goodness engineering into the 30-year-old discipline of genetic engineering.
Elaborate Lego creations, skyscrapers, and integrated circuits all have one fundamental feature in common: They are built from a multitude of simple constituent parts, which interact with one another in predictable ways.
Take electronics. Cellphones, iPads, and similar gadgets contain many millions of individual components, some highly specialized, some more generic. The latter category includes their many resistors, capacitors, and transistors. To reduce them to their bare essence, these generic parts take an input signal, transform it, and spit out a result. The same elements are used over and over for all sorts of things, their function being a product of their particular arrangement.
Curiously enough, the genetic machinery inside a cell works in much the same manner. It takes certain input signals—the concentration of one or more kinds of molecules—processes them in some fashion, and produces an output signal, the concentration of yet another kind of molecule. The problem for genetic engineers is to create the right processing “circuitry” to get a particular job done. This is tougher than in electrical engineering because the necessary parts have generally had to be handcrafted and assembled from scratch each time by teams of Ph.D.s working at the cutting edge of molecular biology. Someone trying to engineer a cell to do something new can’t just mix and match standard components from a library of well-characterized parts the way electrical engineers generally can.
We and other synthetic biologists hope to change that. If we are successful, genetic engineers will one day be able to draw on a set of industry-standard tools for design and simulation. They will then combine widely available and well-characterized components to fashion organisms that produce valuable molecules.
So just how would you do that? To start with, you need hardy and prolific cells, typically bacteria or yeast, to serve as biological factories. These cells are easy enough to come by. But in most cases, you won’t get them to crank out the molecule you want without first manipulating the machinery inside them. It’s their genes that determine the nature of the molecules they make, so their genes are what you need to alter.
As you may recall from high school biology class, a gene is a segment of DNA containing a set of instructions for producing something (typically a protein) along with other bits and pieces that help control when that particular something gets made. When biologists talk about expressing a gene, they’re referring to the many things that have to happen for those genetic instructions to be carried out. Cells use sophisticated means to control their genes, turning individual ones on and off as needed. They have to be sophisticated, because the process of switching a gene on is complicated.
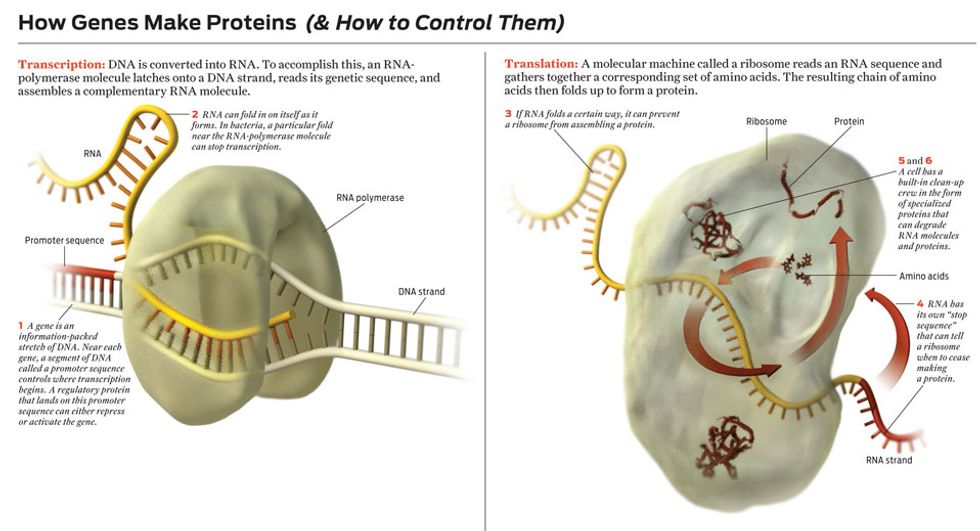
First, a small section of the double-stranded DNA spiral must be made to peel open, exposing the gene’s sequence of nucleotide bases—the alphabet soup of As, Cs, Gs, and Ts you probably remember from your school days. This enables the gene’s DNA sequence to be converted into RNA, a close molecular cousin of DNA. This RNA strand is said to be complementary to the original DNA strand because the new RNA’s As are matched with the original DNA’s Ts; its Cs are matched with Gs. (In truth, RNA uses a nucleic acid denoted by the letter U to match up with DNA’s A, but that’s a minor complication.) Because DNA and RNA share essentially the same nucleic-acid language, biologists call the process of converting DNA into RNA transcription.
Some of these RNA molecules are the end product of the gene, serving the cell with a unique function. But most are just chemical messengers, which carry instructions for the later construction of proteins. So usually the next step is to convert the genetic sequence now coded in messenger RNA into a string of amino acids, the building blocks of proteins. Because there are only 5 nucleotide bases and about 20 amino acids in cells, the language of DNA and RNA is fundamentally different from the language of proteins, which is why biologists call this next stage translation.
Cells use certain molecular clues to tell them when to begin and end the transcription of particular genes into messenger RNA. Other molecules can trigger the breakdown of RNA after it’s made. That creates three potential points of control. Particular molecular cues likewise start and stop translation. Other molecules can degrade proteins. That gives three more, for a total of six distinct knobs for adjusting how a gene is expressed.
The special biomolecules that twiddle these six knobs in the cell are called regulators. With the right ones turned up full blast, a cell will start making a particular kind of RNA and possibly protein continually, basically generating a steady current of this cellular stuff. But the buildup of a different kind of regulator molecule, called a repressor, could then halt the process, acting like a switch and cutting off that current. If this repressor is later removed, a steady current of protein molecules can once again flow into existence.
In electronics, currents usually travel through channels of solid metal or semiconductor, in a direction dictated by the directional push of an electric field. In the cellular world, molecules aren’t so neatly localized. For the most part, they just diffuse through the watery cell interior.
Electronic circuits and cells differ in an even more fundamental way. Consider the transistors on a chip. As long as they are located a modest distance apart, they don’t affect one another. So you can easily wire them together to construct logic gates and other important circuits. A group of transistors in an AND gate will thus function without affecting a different cluster of identical transistors forming an OR gate, for example. To electrical engineers, this degree of independence is so basic they take it for granted.
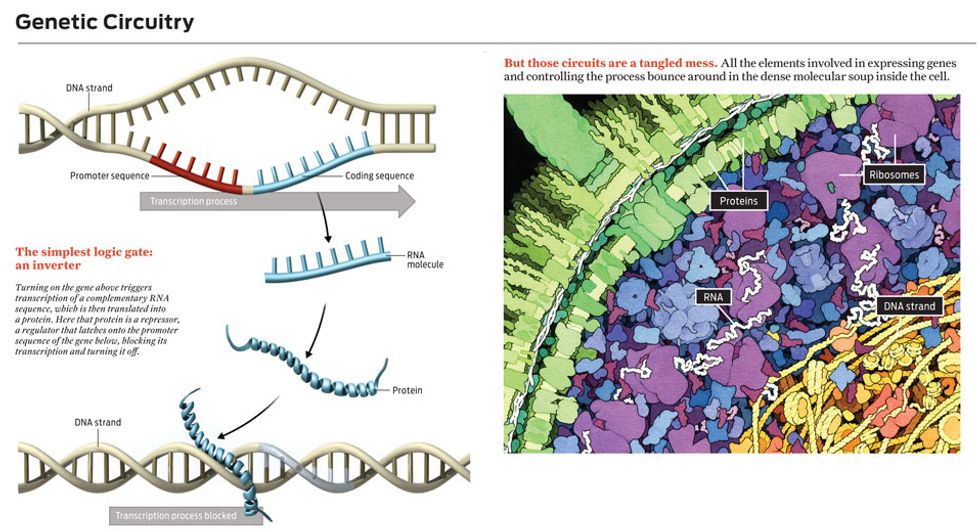
Molecular regulators are more difficult to work with. Let’s say you want to use one to control the creation of a particular kind of messenger RNA. Transcription of a DNA sequence into that RNA starts when a molecule called RNA polymerase binds to a special region of the DNA upstream of the genetic sequence to be transcribed. This special chunk of DNA is called a promoter sequence. After an RNA-polymerase molecule lands on a promoter sequence, it marches along the DNA strand, grabbing nucleotides floating nearby and assembling a complementary RNA sequence.
The simplest way to block this process is to prevent the RNA-polymerase molecule from moving along the DNA strand. Indeed, cells make lots of repressor proteins that bind to DNA near promoter sequences and simply sit there, preventing the movement of RNA polymerase. Such a molecule makes a perfect switch—except that it’s floating around the cell in the presence of millions of other molecules. Even if the regulator molecule recognizes perfectly where it is supposed to latch onto the DNA, it still must find its target, which can take several minutes. How would you like to have to work with switches, transistorized or otherwise, that take minutes to turn off? Cells can’t tolerate such delays either, so they use many copies of a repressor molecule, increasing the probability that at any given time one of them is bound to the target, turning off the gene.
This creates a problem for genetic engineers. Let’s say we want to turn off one gene right now and another gene later on. We can’t just splice in the target sequence for a repressor “switch” near each gene. The presence of multiple repressor molecules in the cell would turn off both. To control them independently, we’d need to use different switches—regulator molecules that work with different target sequences. Nature has devised thousands of different regulator-target pairs over billions of years of evolution. Unfortunately, most of those are very difficult to apply more generally. Synthetic biologists have, however, used some of them as building blocks for their genetic circuits and have made some intriguing biological devices with them.
For example, by engineering DNA so that two different repressor-target sequences sit side by side on a single gene, bioengineers have created a situation where that gene is expressed only when neither regulator—that is, neither input signal—is present. This is the biological equivalent of a NOR gate: If one signal or the other is present at the input, the output is not on.
You can also wire up regulators so that different genes become interdependent, the presence of one gene product controlling a second gene and so on. If you arrange those regulators—some of which repress genes and some of which activate them—so that there is feedback, you can make genetic circuits that work like flip-flops or even as oscillators, where the concentration of some molecule ebbs and flows in a regular way.
These genetic circuits were some of the early successes of synthetic biology. But their functioning is rudimentary. In the future, synthetic biologists will be able to construct far more interesting genetic wetware. Trying to do so using only the natural regulators known to biologists would, however, be like trying to build sophisticated electronic circuits from a grab bag of unlabeled parts, where you’d have to ponder over and account for the poorly documented characteristics of every single component.
For this reason, we’ve decided to build our own regulators. For each of the six gene-expression knobs, we want regulators that differ in the target sequences (or target molecules) they work on but are otherwise identical. To construct them, we’ve identified promising natural molecules to use as starting points. With these molecules, we hope to come up with a whole family of standardized, independently acting components that genetic engineers can use over and over again.
While other synthetic biologists have focused their efforts on manipulating proteins, we have concentrated on engineering RNA. Far from being just an intermediary in gene expression, RNA can adjust five of the six knobs of gene expression, with its ability to start and stop transcription, start and stop translation, and in some cases trigger its own degradation. So it’s a very powerful molecule. Also, from an engineering perspective, RNA is much simpler than protein to work with because its alphabet consists of only four letters, meaning there are fewer variables. Biologists also have a better grasp of how RNA molecules fold into the complex shapes that define their function.
For a long time, synthetic biologists focused on protein repressors to make genetic circuits. The problem is that each of these protein repressors comes from a messenger RNA molecule, which adds an extra middleman in the flow of the circuit. But recently we found an RNA regulator that controls its own transcription based on whether or not another RNA molecule is present, allowing us to build circuits without proteins. So now we can operate solely with RNA. Only a couple of such molecules exist in nature, but by introducing changes in their nucleotide sequences, we’ve created several versions that don’t affect one another’s actions. We’ve also shown that these regulators can be combined in one way to create genetic logic gates and in another way to propagate the signals in genetic circuits, reminiscent of the electrical transistors that inspired our goals.
Our work, and that going on in other synthetic biology laboratories, will slowly build up families of regulators for adjusting the six knobs of gene expression. These families, accompanied by a set of design principles for how to wire them together (figuratively speaking), will eventually give bioengineers the means to manipulate gene expression with great precision and flexibility.
It’s likely that the work synthetic biologists are doing will at first simply add to our collective grasp of the underlying science. And that’s okay. But we’re optimistic that given enough time, we will be able to create and control complex genetic circuits that function as designed. After that, we can start to tackle other challenges, such as constructing more specialized components to accompany our transistor-like building blocks. At that point, genetic engineers will be well positioned to solve some of the world’s biggest problems—including those of food production, land use, pollution, and health. That’s because we’ll finally be using standardized parts and design tools, just as other kinds of engineers have been doing for a great many years.
This article originally appeared in print as “The Hunt for the Biological Transistor.”
About the Authors
Julius B. Lucks and Adam P. Arkin write about how to make genetic engineering more like electrical engineering. Arkin is a professor of bioengineering at the University of California, Berkeley. Lucks, currently a postdoctoral fellow at UC Berkeley, will be joining the chemical engineering faculty at Cornell. Although trained in chemistry, Lucks has done plenty of Python coding, and Arkin’s hacking goes all the way “back to the days when they had Heathkit stores and Radio Shack had stuff you could actually use,” he says.