23 May 2012—Bioengineers looking to turn microbes into manufacturers have longed for a kit of components as regular and predictable as those used by electrical engineers. But biology is a lot messier. Now a group of engineers at Stanford University say they’ve managed to make one such component—the genetic equivalent of a reliable memory device. In a report published this week in Proceedings of the National Academy of Sciences, they detail how they developed rewritable DNA memory that works in living cells and can keep its data even as cells divide and multiply. DNA memory already exists but has been limited to write-once versions that can record only as many cellular events (such as cellular divisions) as there are bits. But the reversible storage system the Stanford researchers have ginned up is capable of being expanded to record a potentially huge number of events—2n events, where n is the number of bits.
In the short term, say its inventors, this improved biological memory could enhance the study of how we age and how cancer grows. But much further out lies the possibility of reprogramming cells to slow the aging process or to act as sentries that prevent cancer’s uncontrolled cell division.
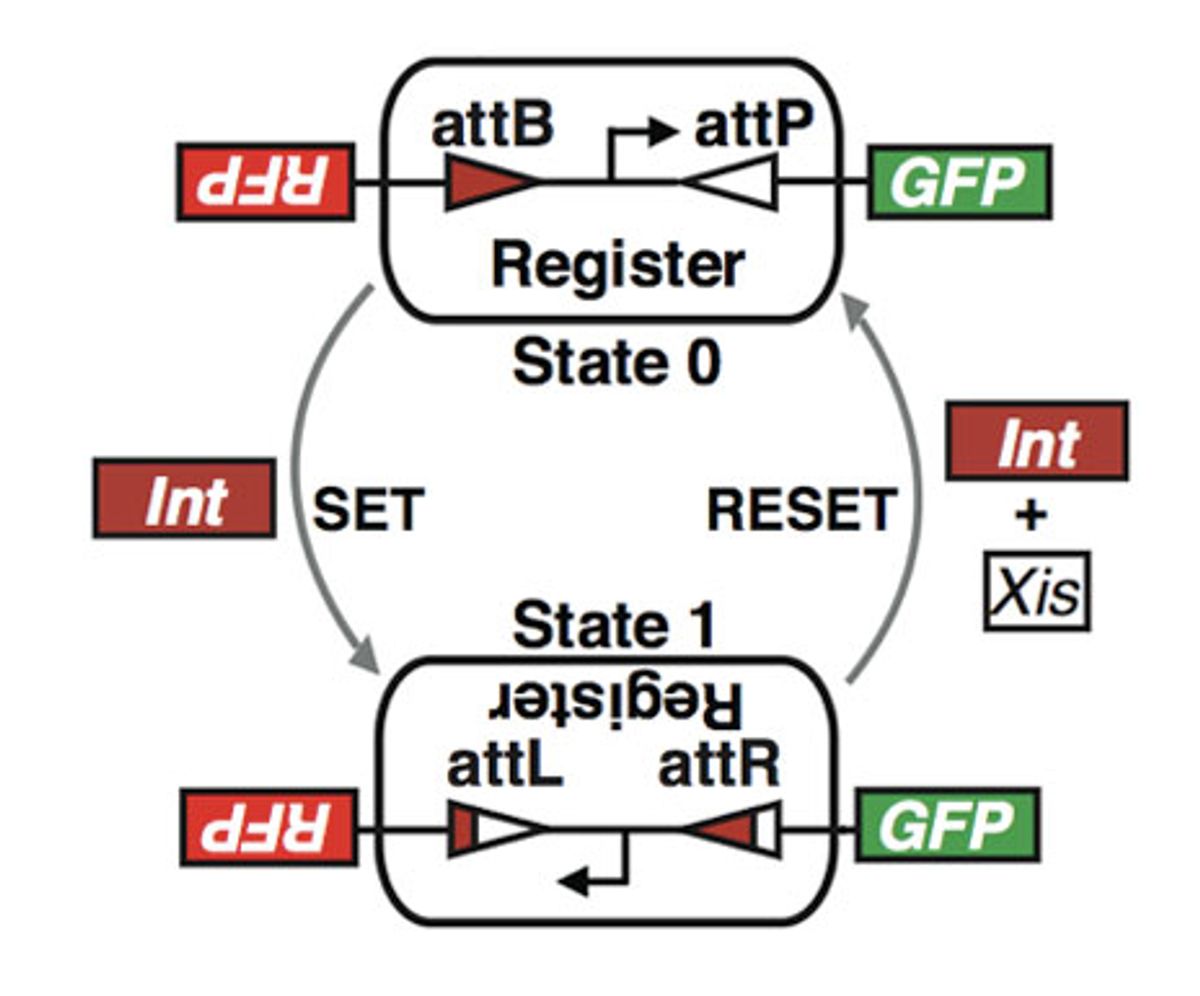
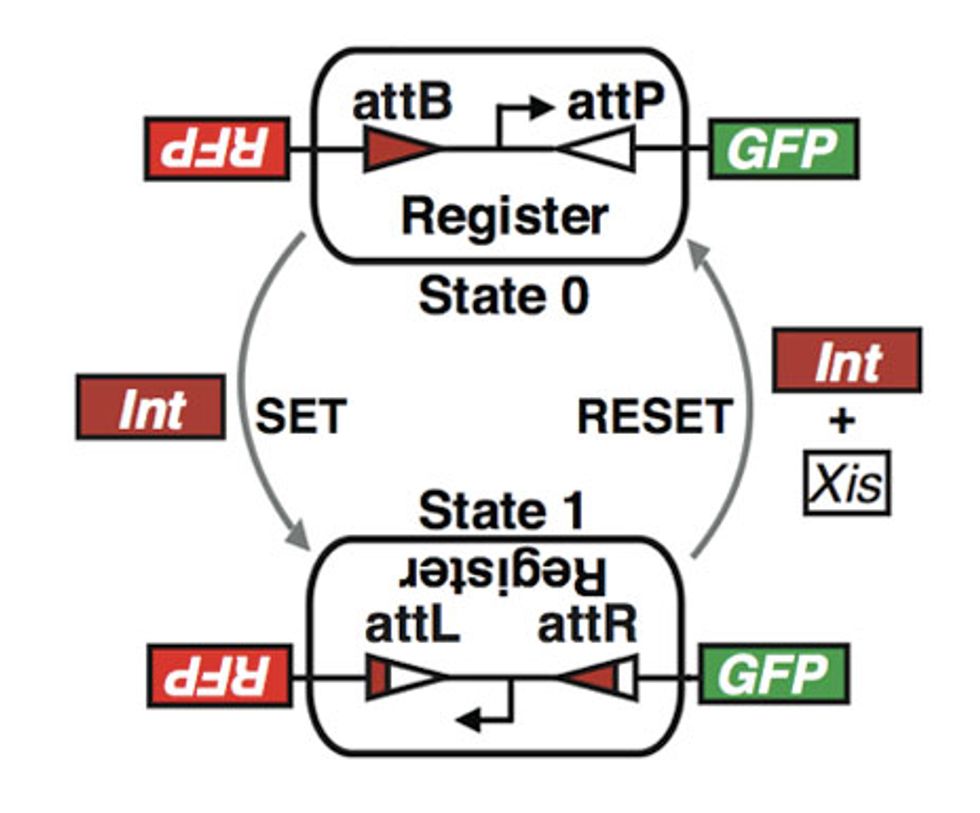
The rewritable recombinase addressable data (RAD) module created by the Stanford bioengineers is a segment of DNA that switches between states when the bacteria carrying it encounters specific proteins. A class of proteins called “integrases” scan DNA sequences until they find two specific sequences (attachment sites called attB and attP) and bind to them. The integrase then cuts out the DNA strand between those sites, flips it over, and reattaches it so that string of base pairs reads in reverse. This chemical process, which the researchers refer to as “setting,” also changes the characteristics of the attachment sites attB and attP. (They become attL and attR, respectively.) This upside-down state, says the Stanford team, is the equivalent of a 1 in an electronic memory device.
Adding integrase mixed with another class of protein called “excisionases” reverses the process, “resetting” the DNA strand to its original, or 0, state. (Excisionase alone has no effect that anyone is aware of.) Drew Endy, a Stanford assistant professor of bioengineering, who led the research, says that the major technical hurdle the group had to overcome was avoiding what’s called “bidirectionality.” That is the tendency for some recombinase proteins (the respective versions of integrase and excisionase that the researchers chose are two of many) to cause the RAD module to flip, and then to cause it to flip back to its previous state before the change in state is recorded.
But in the end, say the researchers, they created a system of DNA registers that switch when, and only when, they’re in the presence of the protein-based inducers. As important, they note, is that the states can be switched repeatedly with no performance degradation.
“Developing biological systems, especially those based on DNA and cells, that ‘compute’ like digital computers has been challenging,” says Steven Benner, a distinguished fellow at the Foundation for Applied Molecular Evolution in Gainesville, Fla. Benner explains the nature of the challenge, noting that “biological molecules, like all molecules, intrinsically do ‘analog’ computation better than ‘digital.’ [The Stanford researchers'] latest work is a big step toward getting digital behavior from structures that are, fundamentally, not digital.”
Asked how much data the device they demonstrated is able to store, Endy proudly reports that it is currently capable of storing 1 bit, as in roughly a hundred billionth of the amount of data that can be stored on a key-fob-size USB flash drive. Though the DNA memory device’s capacity is relatively minuscule, “its purpose is not to compete with silicon, but to get access to data storage in places where silicon doesn’t work,” says Endy.
In fact, says the Stanford researcher, 8 bits is more than enough to keep track of changes in any replicating biological system. With that capacity, he envisions applications such as a fail-safe element in cellular therapeutics. When, say, a cancer patient is injected with living cells reengineered to attack a tumor, the RAD module could be set to control the rate and number of cell divisions so that the cure doesn’t morph into a curse.