What AI Can–and Can’t–Do in the Race for a Coronavirus Vaccine
The precious few molecules that could end the COVID pandemic are hidden by millions that can't. Can AI help find them in time?
In an achievement that would have startled biomedical researchers merely a year ago, vaccines against COVID-19 were already being tested in humans this past March, less than three months after the initial outbreak was identified in China. Many of those vaccines owed their speedy start to the power of artificial intelligence (AI).
The feat is a promising and remarkable turn in the 200-year-plus history of immunization. The experience may revolutionize the way vaccines are created, potentially saving countless lives in epidemics yet to come.
As of early September, there were 34 vaccine candidates being tested in humans, according to the World Health Organization (WHO). Another 145 candidates were being tested in animals or in the lab, says WHO, which keeps a running worldwide list. Those are astonishing numbers, considering that less than a year ago no one had heard of the novel coronavirus, now known as SARS-CoV-2, which causes the respiratory disease COVID-19. It typically takes many years, or even decades, to develop a vaccine; until now, the speed record was held by the mumps vaccine, which went from a collected sample to a marketed product in about four years.
It's no wonder that research is sprinting ahead. Our societies and economies likely won't return to normal until a highly effective vaccine has been administered to a substantial portion of the planet's population. The search for a vaccine is now a vast undertaking, involving thousands of researchers at hundreds of laboratories around the world spending billions of dollars. It's like a moon shot in its magnitude, ambition, and intensity.
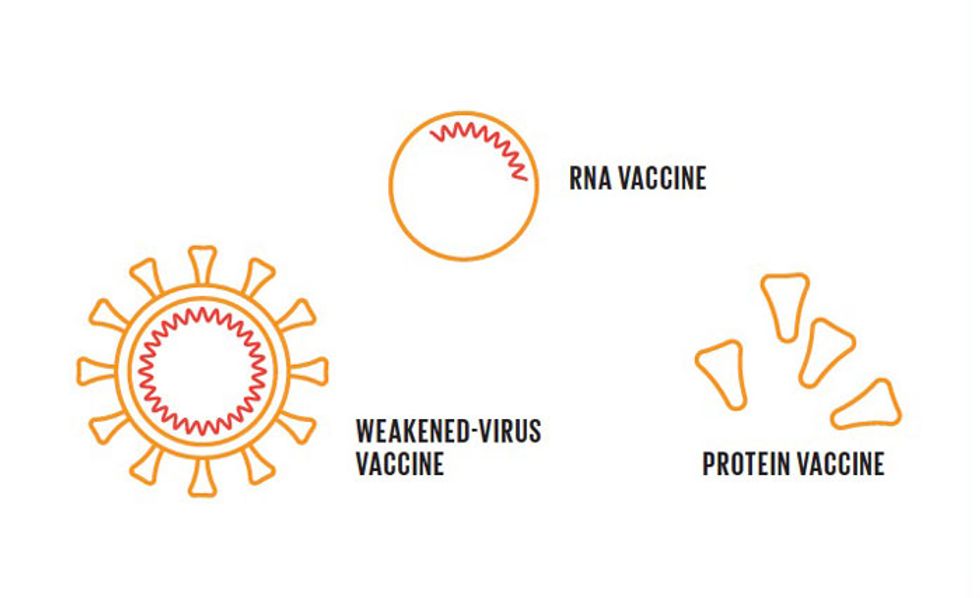
Laboratories are pursuing at least eight different types of vaccine. These include traditional ones based on inactivated viruses, as well as new, more experimental ones involving the use of genetic material—so-called DNA and RNA vaccines—as well as others based on special proteins or other biological agents.
At stake are not only human lives but also a piece of a global vaccine market that was estimated at US $35 billion even before COVID-19. Governments, philanthropies, and pharmaceutical companies have been spending accordingly. In July, the U.S. government agreed to pay pharmaceuticals giant Pfizer and German biotech firm BioNTech nearly $2 billion for 100 million doses of a vaccine, if and when it becomes available. Other major vaccine initiatives worldwide also have funding in the 10 figures.
Machine-learning systems and computational analyses have played an important role in the vaccine quest. These tools are helping researchers understand the virus and its structure, and predict which of its components will provoke an immune response—a key step in vaccine design. They can help scientists choose the elements of potential vaccines and make sense of experimental data. They also help scientists track the virus's genetic mutations over time, information that will determine any vaccine's value in the years to come.
“AI is a powerful catalyst," says Suchi Saria, a professor at the Johns Hopkins Whiting School of Engineering who directs the university's machine-learning and health care lab. AI enables scientists “to draw insights by combining data from multiple experimental and real-world sources," she explains. These data sets are often so messy and challenging that scientists historically haven't even attempted these sorts of analyses, she adds.
As AI tools become more powerful, researchers are anticipating a time when computational methods could help scientists solve our most vexing vaccine challenges—such as finding an effective HIV vaccine, or creating a flu vaccine that's good for more than a year.
The excitement surrounding new computational techniques comes with a caveat: AI cannot replace or speed up the most crucial, time-consuming aspect of vaccine development. Animal and human trials must happen via pure human effort, with thousands of scientists, health care workers, and participants logging their experience with a vaccine in real time. “Computation helps you optimize your chances of success, but ultimately you have to roll up your sleeves and do it in the lab," says Jacob Glanville, founding partner at Distributed Bio and its subsidiary, Centivax, which are developing vaccines for flu, HIV, and other pathogens using computational bioengineering.
Still, in the quest for a COVID vaccine, AI has done more than it ever has before. And it is just part of a larger suite of computational tools that are revolutionizing vaccine R&D. Few people may be thinking about the next pandemic, but researchers are already starting to understand how these tools will do quite a bit more the next time around.
Virus vs. Immune System
Illustrations: Chris Philpot The Attack
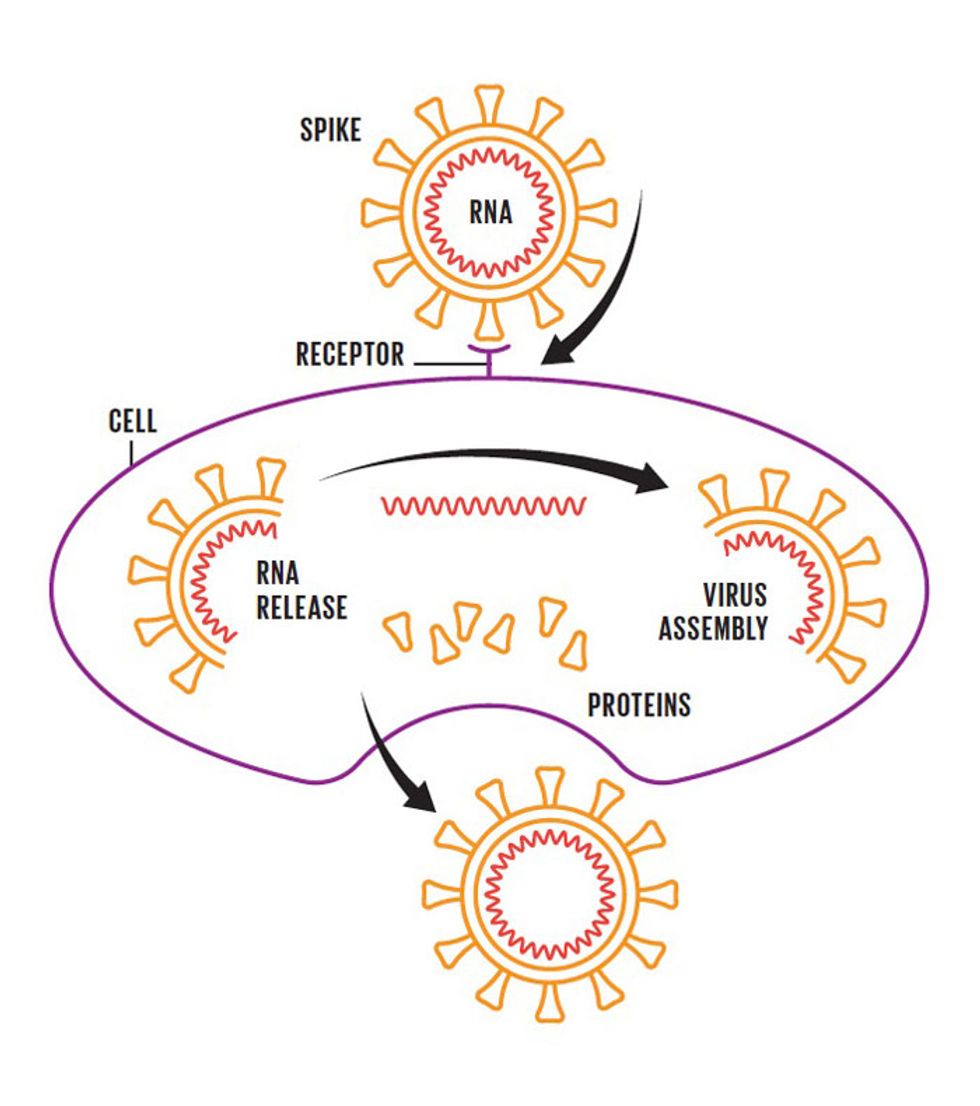
The coronavirus has spikes that bind to receptors on the surface of certain human cells. The virus then fuses with the cellular membrane and releases its RNA genome into the cell. Next, the cell churns out copies of that RNA, as well as the structural proteins needed to assemble new viral particles—which are released into the body.The Defense
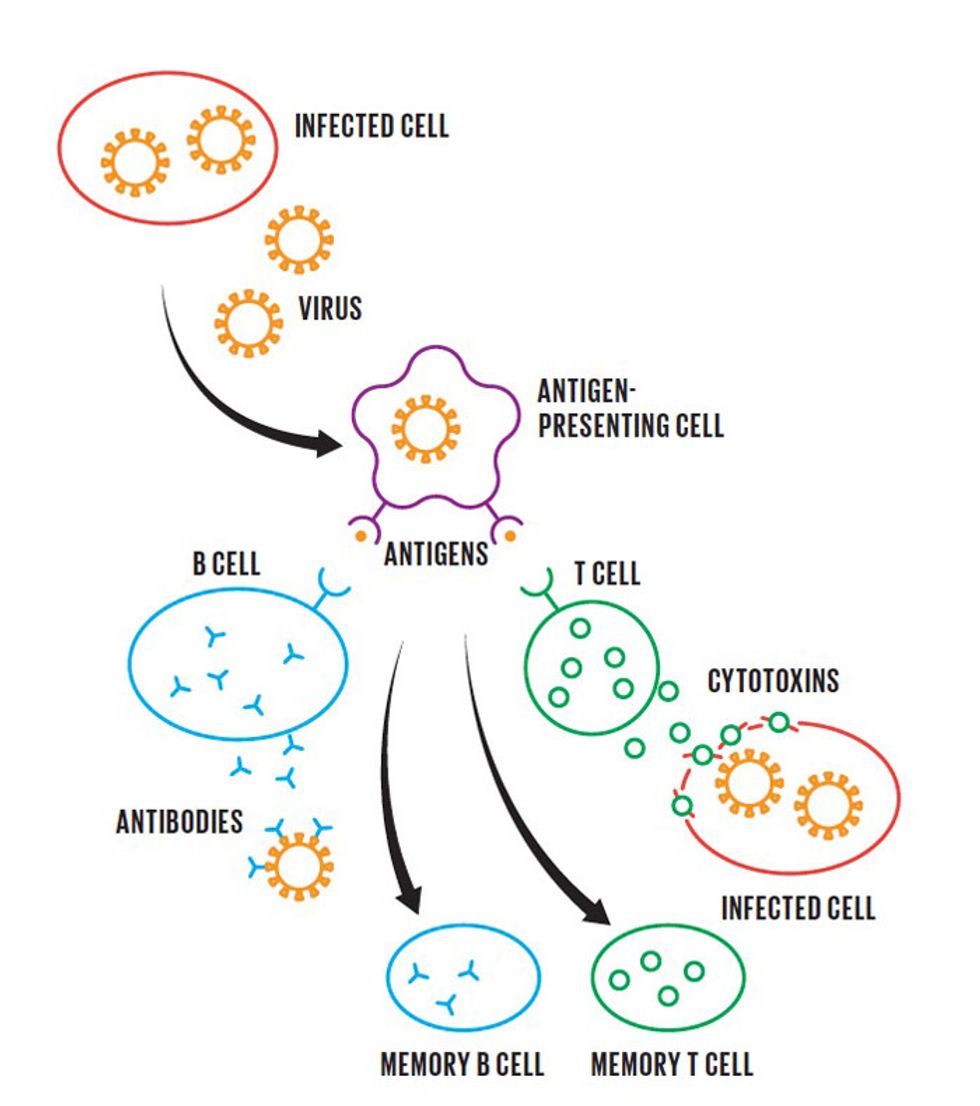
In the body's adaptive immune response, specialized cells engulf the virus and present fragments of it, called antigens, to activate immune cells. B cells begin to make antibodies that bind to viral particles and prevent them from attaching to healthy cells, while T cells destroy cells that have already been infected. Meanwhile, memory B and T cells take note of the antigens, ensuring that the body will respond quickly if it encounters the coronavirus againRaising the Alarm
The task of vaccines is to present antigens that will trigger this immune response without making the person sick. Vaccines come in various forms, including weakened forms of the entire virus, specific fragments of the virus, and either DNA or RNA that causes a cell to make specific fragments of the virus.AI Assistance
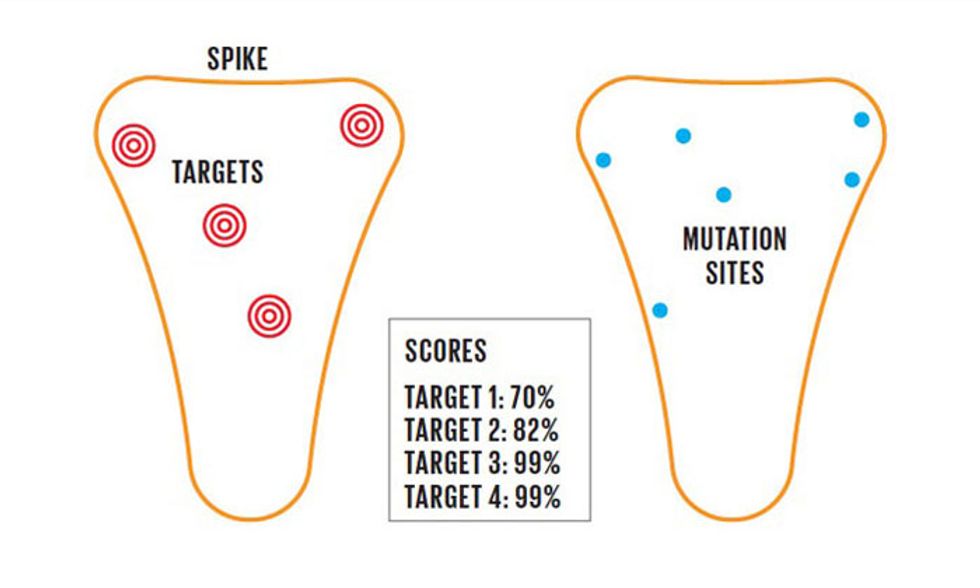
One role for AI in vaccine design is to study the proteins that make up the virus, which include the spike protein. By examining its complex structure, an AI system can sort through thousands of components to identify those that are most likely to trigger a robust immune response. What's more, viruses are always mutating. AI systems need to identify components that are unlikely to mutate, to ensure that a vaccine will remain effective over time.
Modern vaccinedesign is a hugely information-intensive endeavor, starting with the reams of data needed to understand both the virus and our immune system's reaction to it. There are more than 200 viruses known to infect human beings, and each of them is distinct in its mechanisms, behavior, and ultimately, its cures.
Though they vary in the details, viral attacks on the body mostly start off the same way. When a virus gets into the body—say, through the mouth or nose—it infiltrates healthy cells by binding to receptors on the cells' surfaces. The virus can then hijack the cells' machinery to make more copies of itself, and an infection ensues.
Putting a halt to all this is the job of the immune system, which hunts and destroys pathogens such as viruses and bacteria that cause disease. As a first step, the immune system sends a variety of basic weapons to the infection in what's called the innate immune response.
If that's not enough to control the infection—for example, if the pathogen is new to our bodies and the generalized weapons don't work—the immune system's adaptive immune response brings in the bigger guns. Adaptive immunity depends on two types of white blood cells, called B cells and T cells. B cells produce specialized proteins called antibodies, which bind to the pathogen and prevent it from entering healthy cells. T cells, meanwhile, can destroy cells that have been infected by the virus, to keep them from making more copies of it.
It takes days for the adaptive immune response to get revved up enough to begin wiping out a new virus. Our bodies have B cells and T cells tailored for nearly every pathogen the world can throw at us, but it takes time for the right immune cells to find the invader and multiply. In the meantime, we get sick.
The good news is that while this war is raging, the immune system also produces memory B and T cells, which make a record of the battles. If we get exposed to the same pathogen again, the immune system has an arsenal at the ready and responds much more rapidly. We may experience mild symptoms, or none at all.
The goal of a vaccine, then, is to expose the body to a pathogen without making us sick so that the immune system is primed to fight it on any subsequent exposure. This can be done by exposing the body to specific pieces of a virus or weakened versions of it. Crucially, the vaccine must include key parts of the virus, called antigens, that are immunogenic, meaning that they're recognizable to B cells and T cells and will therefore trigger the desired adaptive immune response.
When faced with a new pathogen, the first question for vaccine designers is: Which parts of it are the most immunogenic? A typical virus consists of genetic material, either DNA or RNA, encapsulated by one or more layers of proteins. The outer membrane is often studded with so-called spike proteins, which enable the virus to bind to receptors on a host cell and inject its payload of genetic material. For this reason, spike proteins are a typical target for vaccines. If the immune system creates antibodies that disable the spike protein, the virus cannot break into cells.
However, for any given virus there are tens of thousands of different subcomponents of the outer proteins that the immune system can recognize, and therefore tens of thousands of different possibilities for vaccine targeting. This is a prime opportunity for AI. Machine-learning tools can predict, based on training data sets from known pathogens, which pieces of the virus the immune system is most likely to recognize.
Armed with this information, immunologists can design vaccines around a more manageable number of potential targets. The targets are then integrated into vaccine candidates and tested in animals to see if they provoke a good immune response. Machine learning “gives you a numerical score," says Tayab Waseem, a public policy fellow at the American Association of Immunologists and director of medical informatics and AI integration at Wagner Macula & Retina Center, based in Norfolk, Va. “Everything over a certain score—say, 99 percent—I'll be willing to go into the lab to test out."
In the early weeks of the pandemic, a team of computer scientists led by Russ Altman and Binbin Chen, at the Stanford Institute for Human-Centered Artificial Intelligence (HAI), used machine learning to do just that. Using the neural-network algorithms NetMHCpan-4.0 and MARIA, and a linear-regression model called DiscoTope, the researchers came up with a list of targets on the novel coronavirus that were most likely to provoke an immune response. These targets, or epitopes, are components of the virus that B cells and T cells will likely recognize.
As expected, many of the system's top recommended targets were located on the virus's spike protein. Chen's team recommended, in a paper on the preprint server bioRxiv, that these epitopes be included in the design of COVID-19 vaccines. “We feel pretty confident that we'll get an immune response at the cellular level against what we predicted as targets," says Chen. “But there is a big gap between cellular response and clinical response," he adds.
Chen's machine-learning tools are among several dozen that have been built over the years to aid immunology work. In the past, machine learning has been a “minor sidekick" in vaccine development, according to Chen. But for COVID-19, “people in both academic and industry labs ran more computational studies," he says. “I suspect that all the pharma companies who developed a vaccine also ran a computational analysis."
Having identified a target on the virus's surface, researchers can then develop a vaccine. If the plan is to use an inactivated virus as a vaccine, for example, researchers will grow the live virus in the lab and kill it using heat, radiation, or a chemical method so that it can't replicate when injected into the body. Then researchers must make sure that the key immunogenic components weren't damaged when the virus was killed, as those parts must be intact in order to provoke an immune response. The next steps are to test the vaccine in the lab, then in small animals, and finally in humans.
Select COVID-19 Vaccine-Development Projects
| ALLIANCE | NATIONS | VACCINE TYPE | STATUS (Phase 1, 2, 3, or Approval) | FUNDING, US $ | MAIN FUNDERS |
|---|---|---|---|---|---|
| University of Oxford, AstraZeneca | United Kingdom, United States | Viral-vector (adenovirus) | Phase 3 | $1.25 billion |
U.S. and British governments |
| Sinovac Biotech | China | Inactivated virus | Phase 3 | $15 million | Advantec Capital, Vivo Capital |
| Wuhan Institute of Biological Products, Sinopharm | China | Inactivated virus | Phase 3 | $142 million (with Beijing Institute of Biological Products) | Chinese government |
| Beijing Institute of Biological Products, Sinopharm | China | Inactivated virus | Phase 3 | $142 million (with Wuhan Institute of Biological Products) | Chinese government |
| Moderna, National Institute of Allergy and Infectious Diseases | United States | RNA vaccine (mRNA) | Phase 3 | $2.48 billion | U.S. government |
| Inovio Pharmaceuticals/ International Vaccine Institute | United States | DNA vaccine | Phase 1/2 | $97 million | U.S. government (Department of Defense) and others |
| CureVac | Germany | RNA vaccine (mRNA) | Phase 2 | $440 million | $355 million from German government plus $85 million loan from European Investment Bank |
| BioNTech, Fosun Pharmaceutical, Pfizer | United States, China, Germany | RNA vaccine (mRNA) | Phase 3 | $1.95 billion | U.S. government |
| Gamaleya Research Institute of Epidemiology and Microbiology | Russia | Viral-vector (adenoviruses) | Approved for limited use | NA | Russian government |
| CanSino Biologics, Institute of Biotechnology at the Academy of Military Medical Sciences (China) | China | Viral-vector (adenovirus) | Approved for limited use | NA | Chinese government |
To train software to sift through target sites on a virus, it's important to first understand the three-dimensional structure of viral proteins. Viral proteins are made of linear chains of chemicals called amino acids, which spontaneously fold into compact, ribbonlike structures. Vaccine developers must choose targets on the virus's outer layer that face outward, so that they're physically accessible to immune-system weaponry.
When the pandemic hit, researchers at the University of Basel, in Switzerland, used a protein-modeling tool called Swiss-Model to predict the structures of the proteins on the outer surface of the SARS-CoV-2 virus. Their predictions were later shown to be consistent with the virus's actual protein structures. Similarly, the London-based AI company DeepMind applied its neural network, AlphaFold, to predict the three-dimensional shape of SARS-CoV-2 proteins based on the virus's genetic sequence.
Despite these successes, not all researchers are enthusiastic about the promise of AI for this component of vaccine R&D. They note that, AI or no AI, the spike protein was an obvious target, based on knowledge of other coronaviruses and experimental work with SARS-CoV-2. “There are a lot of methods to identify immunogenic regions of pathogens that do not require artificial intelligence," says Glanville of Distributed Bio. Algorithms that predict such targets are nice to have, he says, but probably not necessary in the case of COVID-19. “AI still has the challenge of proving that it works better than simpler methods," such as serological screening, epitope mapping, and structural biology, he says.
But AI can do much more than zero in on the immunogenic sites on a virus. Many vaccine developers are already using computational tools to design and synthesize the genetic components of DNA-based vaccines. Inovio Pharmaceuticals in San Diego, one of the 34 groups with a COVID-19 vaccine in human trials, is one example.
“The team at Inovio waited enthusiastically for the genetic sequence of the virus to be posted online," says Kate Broderick, senior vice president of R&D at Inovio. “When it was uploaded by the Chinese authorities on January 10, our scientists immediately entered the sequence into our algorithm, and within 3 hours they had a fully designed and optimized DNA medicine vaccine," she says.
Inovio's DNA vaccines work by mimicking a part of the genetic sequence of the pathogen. These so-called nucleic-acid vaccines contain segments of genetic instructions, in the form of DNA or RNA, that code for a key immunogenic component of the virus. When the nucleic acid is inserted into human cells, the cells produce the antigen, which triggers an immune response. Inovio researchers knew, based on previous research on other coronaviruses, that the spike protein of SARS-CoV-2 would likely elicit an immune response. So that region of the virus's genome became their starting point for a vaccine.
There are many different ways to write out a DNA sequence that codes for the production of the same protein. To find the one that will work best as a vaccine, that bit of code has to be enhanced with other genetic and molecular elements. Inovio's proprietary gene-optimization algorithm showed researchers how to do this in such a way that the vaccine would provoke the large-scale production of an immunogenic spike protein.
Inovio's COVID-19 vaccine went from bench to bedside in just 83 days, Broderick says. The vaccine performed well in animals—as shown by a study that she and her colleagues published in May in Nature Communications. In late June, the company announced that the vaccine proved safe and appeared to provoke immune responses in 40 healthy people in a trial in the United States. The vaccine also provided protection for four months in monkeys that were vaccinated and then exposed to the virus, according to a report from Inovio in late July.
Keeping up with a virus's genetic changes also presents a challenge well-suited for computational analysis. Viruses are constantly mutating in small ways, so a vaccine must be designed around a relatively stable region of the virus's genome—a region of its genetic code that doesn't tend to mutate. “There are certain parts of the surface proteins on the virus that have a very high turnover, which you only find out as you sequence it and get changes in structure as it mutates," says Waseem of the Wagner Macula & Retina Center.
Over the last 10 months, tens of thousands of COVID-19 virus samples taken from patients around the world have been genetically sequenced and uploaded into an online repository hosted by the Global Initiative on Sharing All Influenza Data (GISAID), in Germany. Algorithms that compare those sequences can reveal which segments of the virus's genome frequently change, and which segments don't. As the virus continues to conquer new territories, researchers will keep tabs on their ever-changing foe.
All of this work takes a lot of computing power. In March, the White House announced that it would collaborate with public and private groups to provide researchers worldwide with access to the most powerful supercomputers in an effort to “rapidly advance scientific research for treatments and a vaccine."
Called the COVID-19 High Performance Computing Consortium, the program includes resources from the U.S. Department of Energy National Laboratories and several universities and private companies, such as IBM and Hewlett-Packard Enterprise. The consortium boasts nearly 80 active projects, which have access to over 400 petaflops of computing power.
Mahmoud Moradi, a computational chemist at the University of Arkansas, in Fayetteville, led one of those projects. He used supercomputers at the Texas Advanced Computing Center to create enhanced 3D simulations of coronavirus spike proteins. The simulations revealed that the spike proteins become active and infect human cells much faster than those of a previous infectious coronavirus, SARS-CoV-1, which caused an outbreak in Asia in 2003.
Broderick at Inovio says that this kind of research is vital to vaccine-development teams. “Scientists can learn a huge amount of relevant information to assist with vaccine design," she says, “as well as understanding the mechanisms behind the pathogenesis of this virus."
Once a vaccine candidate is designed, the bulk of the work then shifts to testing. Vaccines are first tested in the lab on cells and on animals, and then on increasing numbers of people in clinical tests. Tens of thousands of trial volunteers will have received a vaccine before it's approved by U.S. regulators.
Unfortunately, AI tools can't replace those time-consuming steps. They might be able to predict which antigens the immune system will see, but what the immune system will actually do, in a live human, is beyond the capabilities of today's computers. “The human body is so complex that our models cannot necessarily predict with reliability what this molecule or this vaccine will do for the body," says Oren Etzioni, CEO at the Allen Institute for Artificial Intelligence. “That's why we have these slow and painful trials—our predictive models aren't good enough to give you reliable data."
Although AI can't predict the success of human trials, it can make sense of the mountains of data from these experiments by looking at all the parameters and finding patterns that a human brain might not spot. As vaccine candidates advance to second and third phases of clinical testing, thousands of patients will be involved, and AI systems will be key in rapidly analyzing the clinical and immunological data.
And as more researchers add their studies to the ever-increasing body of literature on the novel coronavirus, scientists will need help sorting through those papers. The Allen Institute developed a resource called CORD-19 that provides more than 130,000 scholarly articles on COVID-19 in machine-readable format. The Kaggle community, among other groups, leveraged the data set to create multiple AI systems to help researchers keep up with literature and answer high-priority research questions.
“I believe that within a decade AI will be an indispensable part of any medical researcher's tool kit both for scouring the literature and for analyzing experimental data," says Etzioni. And when the next pandemic comes—because there will always be a next pandemic—researchers will be poised to unlock the secrets of the deadly pathogen, design many potential vaccines to protect us, and rapidly identify the ones that can prevent a disaster like COVID-19 from befalling humanity again.
This article appears in the October 2020 print issue as “AI Takes Its Best Shot."