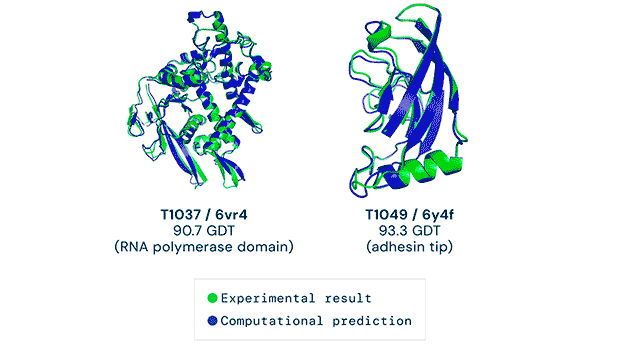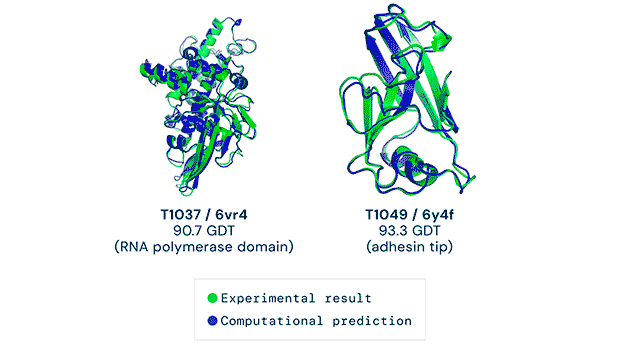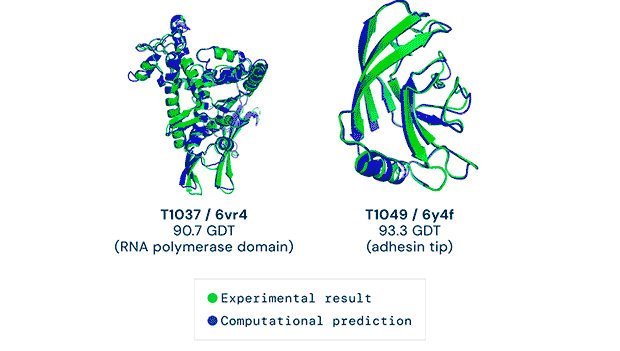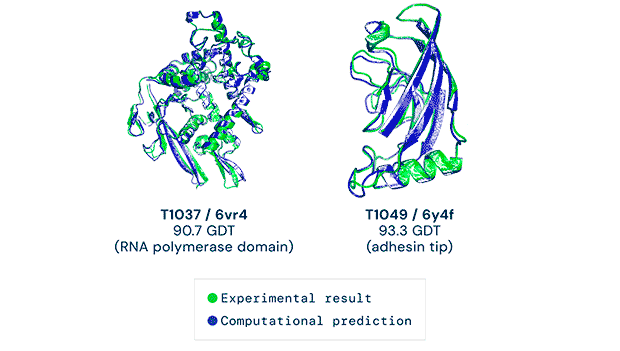
Any successful implementation of artificial intelligence hinges on asking the right questions in the right way. That’s what the British AI company DeepMind (a subsidiary of Alphabet) accomplished when it used its neural network to tackle one of biology’s grand challenges, the protein-folding problem. Its neural net, known as AlphaFold, was able to predict the 3D structures of proteins based on their amino acid sequences with unprecedented accuracy.
AlphaFold’s predictions at the 14th Critical Assessment of protein Structure Prediction (CASP14) were accurate to within an atom’s width for most of the proteins. The competition consisted of blindly predicting the structure of proteins that have only recently been experimentally determined—with some still awaiting determination.
Called the building blocks of life, proteins consist of 20 different amino acids in various combinations and sequences. A protein's biological function is tied to its 3D structure. Therefore, knowledge of the final folded shape is essential to understanding how a specific protein works—such as how they interact with other biomolecules, how they may be controlled or modified, and so on. “Being able to predict structure from sequence is the first real step towards protein design,” says Janet M. Thornton, director emeritus of the European Bioinformatics Institute. It also has enormous benefits in understanding disease-causing pathogens. For instance, at the moment only about 18 of the 26 proteins in the SARS-CoV-2 virus are known.
Predicting a protein’s 3D structure is a computational nightmare. In 1969 Cyrus Levinthal estimated that there are 10300 possible conformational combinations for a single protein, which would take longer than the age of the known universe to evaluate by brute force calculation. AlphaFold can do it in a few days.
As scientific breakthroughs go, AlphaFold’s discovery is right up there with the likes of James Watson and Francis Crick’s DNA double-helix model, or, more recently, Jennifer Doudna and Emmanuelle Charpentier’s CRISPR-Cas9 genome editing technique.
How did a team that just a few years ago was teaching an AI to master a 3,000-year-old game end up training one to answer a question plaguing biologists for five decades? That, says Briana Brownell, data scientist and founder of the AI company PureStrategy, is the beauty of artificial intelligence: The same kind of algorithm can be used for very different things.
“Whenever you have a problem that you want to solve with AI,” she says, “you need to figure out how to get the right data into the model—and then the right sort of output that you can translate back into the real world.”
DeepMind’s success, she says, wasn’t so much a function of picking the right neural nets but rather “how they set up the problem in a sophisticated enough way that the neural network-based modeling [could] actually answer the question.”
AlphaFold showed promise in 2018, when DeepMind introduced a previous iteration of their AI at CASP13, achieving the highest accuracy among all participants. The team had trained its to model target shapes from scratch, without using previously solved proteins as templates.
For 2020 they deployed new deep learning architectures into the AI, using an attention-based model that was trained end-to-end. Attention in a deep learning network refers to a component that manages and quantifies the interdependence between the input and output elements, as well as between the input elements themselves.
The system was trained on public datasets of the approximately 170,000 known experimental protein structures in addition to databases with protein sequences of unknown structures.
“If you look at the difference between their entry two years ago and this one, the structure of the AI system was different,” says Brownell. “This time, they’ve figured out how to translate the real world into data … [and] created an output that could be translated back into the real world.”
Like any AI system, AlphaFold may need to contend with biases in the training data. For instance, Brownell says, AlphaFold is using available information about protein structure that has been measured in other ways. However, there are also many proteins with as yet unknown 3D structures. Therefore, she says, a bias could conceivably creep in toward those kinds of proteins that we have more structural data for.
Thornton says it’s difficult to predict how long it will take for AlphaFold’s breakthrough to translate into real-world applications.
“We only have experimental structures for about 10 per cent of the 20,000 proteins [in] the human body,” she says. “A powerful AI model could unveil the structures of the other 90 per cent.”
Apart from increasing our understanding of human biology and health, she adds, “it is the first real step toward… building proteins that fulfill a specific function. From protein therapeutics to biofuels or enzymes that eat plastic, the possibilities are endless.”
This article appears in the February 2021 print issue as “How AI Conquered Protein Folding.”
Payal Dhar (she/they) is a freelance journalist on science, technology, and society. They write about AI, cybersecurity, surveillance, space, online communities, games, and any shiny new technology that catches their eye. You can find and DM Payal on Twitter (@payaldhar).